7.85: The Hallmarks of Cancer#
- 7.85: The Hallmarks of Cancer
- Facts and numbers
- Abbreviations
- Conventions
- Definitions
- List of viruses
- List of genes
- 1. Cancer the disease
- 2. Molecular cancer introduction: Oncogenes
- 3. Molecular cancer introduction: Tumor suppressor genes (TSGs)
- 3L. Oncogenes; Introduction to Tumor Suppressor Genes
- 4. Familial cancer syndromes; genetic and non-genetic risk factors
- 5. Shaping the cancer genome @weinberg2e/12
- 6. Characterizing the cancer genome
- 7. Functional genomics
- 8. Epigenetic and cancer development
- 9. Signal transduction (I)
- 10. Signal transduction (II)
- 11. Cell cycle @weinberg2e/8
- 12, 13. Metabolism in cancer
- 14, 15. Cell death, senescence, and cancer @weinberg2e/9
- 16. Stem cells and cancer stem cells
- 17. The tumor microenvironment @weinberg2e/13
- 18. Metastasis @weinberg2e/14
- 19, 20. Cancer immunology and immunotherapy @weinberg2e/15
- 21, 22. Principles of cancer therapy @weinberg2e/16
- L21, L22
- 23. Therapy and resistance; early detection and prevention
- @weinberg2e/10 Eternal life: cell immortalization and tumorigenesis
- @weinberg2e/11 Multi-step tumorigenesis
Facts and numbers#
- RBCs have an average lifetime of 120 days (4 months)
- epithelial colon cells live for 5 to 7 days before apoptosis
- keratinocytes of skin die within 20 to 30 days of being formed
Abbreviations#
- IL-2. interleukin 2
- growth-stimulating protein
- GM-CSF. granulocyte macrophage colony-stimulating factor
- growth-stimulating protein
- BrdU. bromodeoxyuridine
- thymidine analog
- 3-MC. 3-methylcholanthrene
- a potent carcinogen and mutagen
- SH1, SH2. Src (sarcoma) homology
- protein domain in tyrosine kinases
- NF1. neurofibromatosis type 1
- GSK-3β. glycogen synthase kinase-3
- KEAP1. Kelch-like ECH-associated protein 1
- ARE. antioxidant response element
- DR. dietary restriction
- MIN. microsatellite instability
- PAH. polycyclic aromatic hydrocarbons
- DSB. double stranded (DNA) break
- CYP. cytochrome P450 enzyme
- HCA. heterocyclic amines
- APE. apurinic/apyrimidinic endonuclease
- AID. activation-induced cytidine deaminase
- used in generating somatic hypermutation in Ig genes
- PCNA. proliferating-cell nuclear antigen; localizes to replication forks
- HDR. homology-directed repair; occurs during late S and phase of the cell cycle.
- -SMA. -smooth muscle actin
- EPC, CEP. circulating endothelial progenitor cells
- PLC-. phospholipase C-
- PDE. phosphodiesterase
- PKC. protein kinase C
- PPAR-. peroxisome proliferator-activated receptor-
- RXR. retinoid X receptor
- SERM. selective estrogen receptor modulator
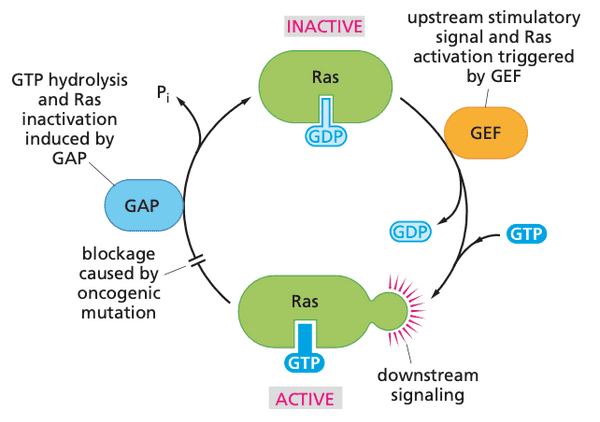
- GEF. guanine nucleotide exchange factor
- GAP. GTPase activating protein
- PH. pleckstrin homology domain; have high affinity for triply-phosphorylated inositol head group
- SMAD. combination of "small" worm phenotype and "mothers against decapentaplegic"
- CKI. CDK inhibitors
- bHLH. basic DNA binding domain + helix-loop-helix
- EpCAM. epithelial cell-adhesion molecules
- DTC. disseminated tumor cell
Conventions#
- nonhuman oncogenes
- gene: uncapitalized three-letter words (e.g. myc)
- gene product: initial capital, roman font (e.g. Myc)
- human genes
- gene: all capitalized, italics (e.g. MYC)
- gene product: all capitalized, roman (e.g. MYC)
Definitions#
- focus. a cluster (e.g., a focus of cells)
- virus stock. a solution of virus particles
- transformation. the conversion of a normal cell into a tumor cell
- contact inhibition (=density inhibition, topoinhibition).
- papilloma. a wart; generally benign lesions that rarely progressed to squamous cell caricnomas of the skin
- papova. a family of viruses signifying papilloma, polyoma, and vacuoles
- T antigens. tumor-associated proteins
- episomes. unintegrated genetic elements
- provirus. the DNA version of a RNA viral genome
- proto-oncogene. a precursor to an active oncogene
- insertional mutagenesis. a mechanism of proto-oncogene activate in which a viral, constitutive promoter takes transcriptional control over a proto-oncogene.
- oncoproteins. retrovirus-encoded oncogene proteins
- hepatoma. liver carcinoma
- endogenous proviruses. proviral DNA that is transcriptionally silent until somehow awakened.
- erythroblastosis. a malignancy of red blood cell precursors
- neuroblastoma. tumors of the peripheral nervous system
- double minutes (dmin). small fragments of extra-chromosomal DNA
- homogeneously staining regions (hsr). regions of the chromosome with uniform stain intensity, indicating copy number gains.
- amplicon. the region of chromosomal DNA that undergoes amplification
- syncytium. a single cell or cytoplasmic mass containing several nuclei, formed by fusion of cells or by division of nuclei
- can be used as an experimental technique to identify dominant vs. recessive phenotypes
- heterokaryon. a cell that contains two or more genetically unlike nuclei
- fusogenic agent. an agent that triggers membrane fusion
- e.g. polyethylene glycol (PEG)
- interstitial deletion. a deletion that occurs from the interior of a chromosome
- null allele. a mutation that results in either no gene product or the absence of function at the phenotypic level
- DNA methyltransferase. enzymes responsible for methylating CpGs
- neurofibromas. benign tumors of the sheaths surrounding PNS nerves
- neurofibrosarcomas (=malignant peripheral nerve sheath tumor, MPNST). malignant tumor of the sheaths surrounding PNS nerves
- gliomas. tumors of the astrocyte lineage in the brain
- pheochromocytomas. tumors arising from the adrenal glands
- variable expressivity. variation in the clinical presentation of a genetic disease, often because of patients' genetic backgrounds
- perineurium. a protective sheath that surrounds a nerve fascicle
- nerve fascicle. a bundle of nerve fibers belonging to a nerve in the PNS.
- blastocoel. the fluid-filled cavity of a blastula
- blastula (=blastocyst in mammals). an animal embryo at the early stage of development with it is a hollow ball of cells.
- morula blastula gastrula
- essential genes. genes required for embryonic development; without them, the embryo dies
- haploinsufficiency. a state in which the presence of only a single functional copy of a gene yields a mutant or partially mutant phenotype
- half isn't always able to recover functionality
- E3 ligase. ubiquitylates proteins and targets them for degradation
- satellite sequences. highly repeated sequences in the genome, often more than 100 nucleotides per repeat unit
- microsatellites. simpler, much shorter sequences of repeats
- transition mutation. when one pyrimidine replaces another (or purine replaces another) (i.e. or )
- transversion. when a purine is replaced by a pyrimidine, or vice versa; the other possible kind of point mutation besides transition (i.e. or )
- peroxisome. organelle involved in the oxidation of various cellular constituents, notably lipids
- abasic site. locations in DNA where no base is present
- hypochlorite ion.
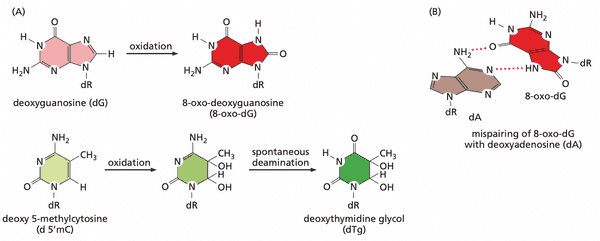
- 8-oxo-dG. a common product of DNA oxidation
- keratose. a benign skin lesion
- alkylating agents. chemicals capable of attaching alkyl groups covalently to DNA bases
- DNA adduct. the chemical entity formed after a carcinogen reacts with a DNA base
- R (restriction) point (=Start, G1/S checkpoint).
- replication stress. unbalanced mitogenic signals cause uncoordinated firing of replication origins and frequent replication fork collapse
- parenchyme. the bulk of functional substance in an animal organ or structure; non-vascular cells.
- anoikis. a form of apoptosis that occurs when anchorage-dependent cells detach from the surrounding ECM.
- serves to excavate lumina when forming ducts in morphogenesis
- bombesin (=gastrin-releasing peptide, GRP)
- bradykinin. promotes inflammation; upon binding to receptor, causes release of arteriole dilators and vein constrictors
- cholecystokinin (=CCK). stimulates release of bile into intestine and secretion of enzymes by pancreas
- neurotensin. regulates luteinizing hormone and prolactin release
- pheochromocytomas. tumors of cells in the adrenal gland
- hemangioblastomas. blood vessel tumors
- pocket proteins. generic name given to pRb and its two cousins p107 and p130.
- diapedesis. the sequence of steps that enable leukocytes to extravasate
List of viruses#
|Name|Type|Virus Family|Approximate size of genome (kb)| |:---|:---|:-----------|:------------------------------| |SV40 (simian virus 40th isolate)/polyoma|DNA|papova|5 | |Epstein-Barr Virus (=human herpesvirus 4, EBV)|DNA|herpesvirus|172| |Kaposi's sarcoma herpesvirus (=KSHV/HHV-8)|DNA|herpesvirus|~165| |murine leukemia virus (MLV)|||| |avian leukosis virus (ALV)|||| |human T-cell leukemia virus (HTLV-I)||||
List of genes#
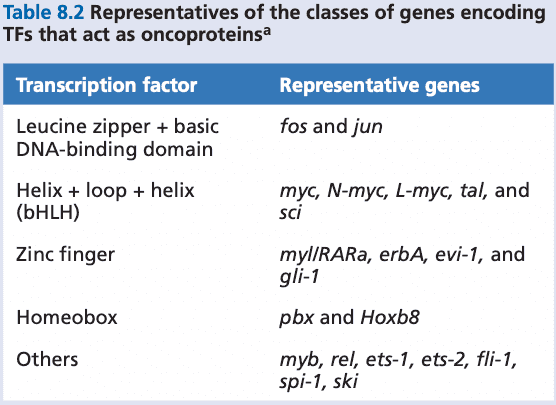
- erbB
- erbB2/neu/HER2
- ras
- humans have three ras genes: H-ras, K-ras, N-ras
- HMGA2. a small non-histone nuclear protein
- capable of functioning as an oncoprotein
- MLL1 (=ALL1). mixed lineage leukemia
- encodes a histone methylase
- PAX3, PAX7. transcription factors
- FKHR. transcription factor
- CDKN2A, CDKN2B. tumor suppressor genes
- inhibitors of CDK4/6
- Wnt. a family of growth factors
- Tcf/Lef. a group of DNA-binding proteins
- MAF. transcription factor family; binds to genes whose promoters include antioxidant response element (ARE) sequences
- MLH1. mutL homology 1
- key protein involved in mismatch repair
- NBS1 (nibrin). involved in initial steps of homology-directed repair.
- TMPR32.
- ERG. Ets-related gene
- p53. implicated in regulation of centrosome number
- CHFR. checkpoint with forkhead and RING finger domains protein
- part of the spindle assembly checkpoint (SAC)
- 14-3-3. traps cycle B-Cdc2 complexes in the cytoplasm, preventing progression into the M phase
- PARP1. poly-(ADP ribose) polymerase 1; recruits repair enzymes for fixing SSBs
- dominant cancer genes
- BRAF
- EGFR
- ERBB2/HER2
- PIK3CA
- IDH1. isocitrate dehydrogenase 1, component of Krebs cycle
- IDH2
- EZH2. histone H2 methylase
- FOXL2. tissue-specific transcription factor
- PPP2R1A
- JAK2
- recessive cancer genes (tumor suppresor genes)
- SETD2. histone H3 methylase
- MLL2. histone H3 methylase
- KDM6A. histone H3 demethylase
- KDM5C. histone H3 demethylase
- PBRM1. part of chromatin restructuring complex
- BAP1. part of chromatin restructuring complex
- ARID1A. part of chromatin restructuring complex
- DAXX. part of chromatin restructuring complex
- ATRX. part of chromatin restructuring complex
- DNMT3A. involved in maintaining cytosine methylation in DNA
- GATA3.
- APOBEC. apolipoprotein B mRNA editing catalytic polypeptide like family
- can bind both RNA and ssDNA
- cytidine deaminase
- SMAD4. critical to TGF- signal transduction
- HIP1. part of complex of proteins that facilitate endocytosis.
- overexpression of HIP1 prevents endocytosis, leading to increased levels of cell surface proteins like EGFR.
- Sevenless. a homolog of the FGF receptor
- Sos. son of Sevenless
- a guanine nucleotide exchange factor (GEF)
- SHP1. phosphatase; responsible for shutting off EPO growht factor receptor
- CDH1. gene encoding the E-cadherin protein
1. Cancer the disease#
- Structure of course
- Molecular drivers of cancer: cancer genes, epigenetics
- Cancer genomics: how does the cancer genome change over time?
- Pathways controlled by cancer genes
- Signal transduction
- Cell cycle (proliferation)
- Cell death
- Stem cell function
- Cancer metabolism
- Tumor microenvironment
- Immune response to cancer
- Cancer treatment, early diagnosis, prevention
2. Molecular cancer introduction: Oncogenes#
- Readings
- Weinberg, pp. 71-130
@weinberg2e/3.1 RSV (Rous sarcoma virus) is discovered to transform infected cells in culture#
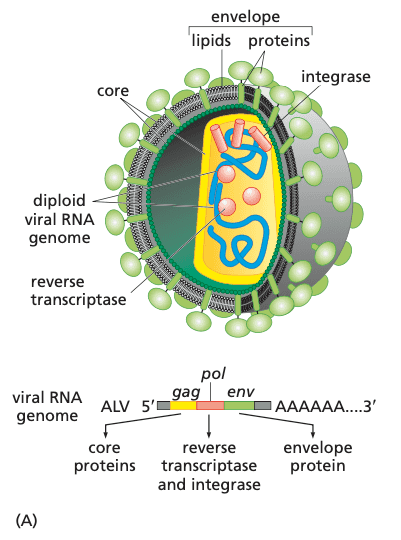
Avian leukosis virus (top) and schematic genome of Rous sarcome virus (bottom). Colors of proteins correspond with the colors of the genes (i.e. env encodes the glycoprotein spikes). The Src protein (pronounced "sarc" for sarcoma, encoded by src gene) in RSV causes cell transformation.
The continued prescence of RSV is needed to maintain transformation#
- Mutant ts (temperature-sensitive) RSV could only grow at 37ºC and not 41ºC.
- 41ºC is the normal temperature at which chicken cells grow.
- Transformed chicken cells with ts RSV:
- If permissive temperature (37): showed cancerous phenotype
- If non-permissive temperature (41): reverted back to normal phenotype
Viruses containing DNA molecules are also able to induce cancer#
- SV40 and human AdV were lytic (and therefore non-tumorigenic) in permissive environments (i.e. human and monkey hosts), but (occasionally) were tumorigenic in non-permissive environments (i.e. rodent hosts).
Tumor viruses induce multiple changes in cell phenotype including acquisition of tumorigenicity#
- Properties of transformed cells
- Altered morphology
- Loss of contact inhibition
- Anchorage independence. ability to grow without attachment to solid substrate
- Immortalization. ability to proliferate indefinitely
- Reduced requirement for mitogenic growth factors in serum
- Increased transport of glucose
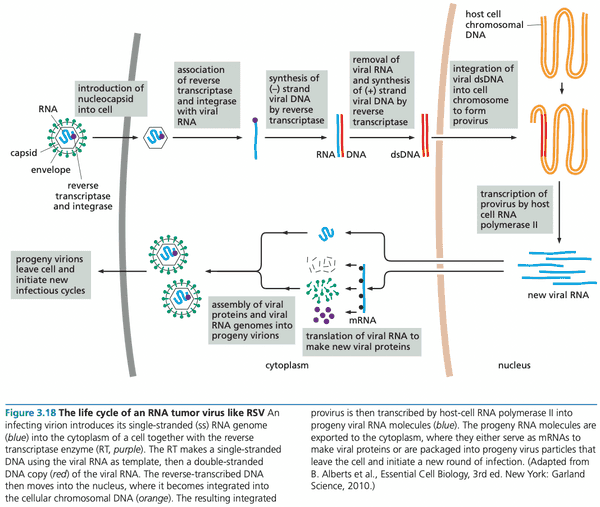
Tumor virus genomes persist in virus-transformed cells by becoming part of host-cell DNA#
- In 1968, it was shown that SV40 viral DNA was tightly associated with chromosomal DNA, indicating integration into the chromosome
- cervical cancer
- HPV
- E2 gene. product represses transcription of viral oncogenes; it is disrupted or discarded when HPV integrates into the chromosome with tumorigenic effect
- E6, E7 oncogenes drive transformation
- HPV
Retroviral genomes become integrated into the chromosomes of infected cells#
- pol gene encodes an integrase along with reverse transcriptase
- because RNA viruses have a dedicated integrase enzyme (in contrast with DNA viruses), they are completely integrated into the host genome, rather than partially (DNA viruses are rarely and randomly integrated into the chromosome).
A version of the src gene carried by RSV is also present in uninfected cells#
- genes for viral replication
- reverse transcription (1 gene: reverse transcriptase + integrase)
- construction of progeny virions (2 genes)
- genes for viral transformation
- very little of genome dedicated to transformation
- hypothesized that a src gene encoded this function
- turned out that src was present in uninfected chicken cells
RSV exploits a kidnapped cellular gene to transform cells#
- RSV likely originally picked up src from one of its hosts
- c-src (cell src) was a proto-oncogene: a precursor to an active oncogene
- v-src lacks C-terminal inhibitory phosphorylation site (tyrosine-527)1
The vertebrate genome carries a large group of proto-oncogenes#
- examples
- v-myc from the MC29 myelocytomatosis virus
- H-ras, K-ras
- more than 30 distincty vertebrate proto-oncogenes have been discovered
Slowly transforming retroviruses activate proto-oncogenes by inserting their genomes adjacent to these cellular genes#
- when a virus integrated next to a proto-oncogene, the constitutive promoter of the virus could activate the proto-oncogene, turning it into an oncogene.
Some retroviruses naturally carry oncogenes#
- HTLV-I doesn't use insertional mutagenesis, nor does it encode mutated cellular oncogenes
- instead, a viral protein called Tax is able to activate transcription of growth-stimulating proteins IL-2 and GM-CSF
- Tax's actual purpose is to activate transcription of proviral DNA sequences, however
@weinberg2e/4.1 Can cancers be triggered by the activation of endogenous retroviruses?#
- tumor viruses are not the only cause of cancer: only two commonly occurring tumor types could clearly be tied to viral, causative agents
- cervical carcinomas
- hepatomas (liver carcinomas)
- could endogenous retroviruses (latent proviral DNA) simply be awakened by non-biological factors?
- no, because many of these sequences have mutated and become nonfunctional.
Transfection of DNA provides a strategy for detecting nonviral oncogenes#
- transfection with calcium phosphate crystals made it possible for cells to take up foreign genetic material
- cells from NIH 3T3 (derived from mouse embryo fibroblasts) were especially good at transfection and chromosomal integration
- still not understood why this works
- needed to choose a cancer cell donor that had highest chance of success
- researchers eventually chose tumors from mouse fibroblasts: C3H10T1/2 mouse cell line treated with 3-methylcholanthrene (3-MC)
- DNA extracted from 3-MC treated mouse fibroblasts was able to transform 3T3 cells
- DNA extracted from healthy mouse fibroblasts (same cell line) were not able to transform 3T3
Oncogenes discovered in human tumor cell lines are related to those carried by transforming retroviruses#
- the same proto-oncogenes activated by viruses could also be activated via non-biological factors
- erbB from avian erythroblastosis virus (AEV) had related gene erbB2/neu/HER2 in human breast cancer
- gene amplification of HER2 correlated with lower survival rates
- make takeaway: a common set of cellular proto-oncogenes might be activated either by retroviruses (in animals) or, alternatively, by nonviral mutational mechanisms operating during the formation of human cancers
Proto-oncogenes can be activated by genetic changes affecting either protein expression or structure#
- mechanisms for proto-oncogene oncogene behavior not obvious
- for retrovirus-associated oncogenes
- regulated promoter constitutive promoter
- for proto-oncogenes activated by non-biological factors
- point mutation (e.g. H-ras oncogene activation: )
G>T- can be regulatory (gene expression) or structural
- point mutation (e.g. H-ras oncogene activation:
- for retrovirus-associated oncogenes
Variations on a theme: the myc oncogene can arise via at least three additional distinct mechanisms#
- three additional mechanisms
- provirus integration
- gene amplification
- chromosomal translocation (e.g. chromosomes 14 and 8 in Burkitt's lymphoma)
- oncogenesis could be due to regulated constitutive promoter
- oncogenesis could also occur because microRNA recognition sites are deleted
- e.g. Let-7 miRNA recognition sites in 3'UTR of HMGA2 mRNA deleted HMGA2 mRNA not degraded increased levels of HMGA2 (still unknown) alters chromatin configuration to facilitate cell transformation
- miRNA also affect cell differentiation; changes in miRNA expression due to chromosomal translocations can prevent cells from differentiating, keeping them in a cell state that easily becomes cancer
- overall, mechanisms leading to overexpression of genes in cancer cells remains poorly understood
A diverse array of structural changes in proteins can also lead to oncogene activation#
- deletions can lead to aberrant protein function
- decapitated EGF receptor constitutively sends signals even when ligand is not bound
- chromosomal translocations can lead to novel hybrid proteins
- chronic myelogenous leukemia: bcr-abl oncogene
- bcr = breakpoint cluster region
- abl = Abelson murine leukemia virus
- fusion protein forms a deregulated Abl protein
- chronic myelogenous leukemia: bcr-abl oncogene
3. Molecular cancer introduction: Tumor suppressor genes (TSGs)#
- Readings
- Weinberg, The Biology of Cancer 3e, Ch. 7
@weinberg3e/7.1 Cell fusion experiments indicate that the cancer phenotype is recessive#
- cell fusion between cancer and normal cells showed offspring that had normal phenotype, indicating that cancer was recessive
- except: tumor viruses that modified the genome had a dominant phenotype
The recessive nature of the cancer cell phenotype requires a genetic explanation#
- hypothesized the existence of TSGs, both of which had to be eliminated for cancer to grow
- mathematically, the chance that both copies of the TSG gene would be damaged seemed too low: .
The retinoblastoma tumor provides a solution to the genetic puzzle of TSGs#
- familial retinoblastoma: likely to have tumors in both eyes, and develop tumors later in life
- sporadic retinoblastoma: single eye, no effect on future tumor development
- Rb is a TSG; if parents passed down a defective Rb gene, then child much more likely to develop retinoblastoma
- sporadic retinoblastoma required two-hits, very unlikely
Incipient cancer cells eliminate wild-type copies of TSGs by a variety of mechanisms#
- mitotic recombination (not meiotic recombination) occurs at rate to per cell generation, much more likely than direct mutational inactivation
- loss of heterozygosity (LOH). a type of genetic abnormality in which one copy of an entire gene and surrounding chromosomal region is lost
- mitotic recombination can be a cause (loss of one parent's allele)
- gene conversion (another potential cause of LOH). DNA polymerase temporarily switches templates and replicates DNA belonging to the homologous chromosome
- chromosomal nondisjunction. chromosomal region breaks off, leading to hemizygosity
The Rb gene often undergoes loss of heterozygosity in tumors#
- test of Rb LOH hypothesis using closely mapped and characterized gene for esterase D
- esterase D served as a surrogate marker
- Rb is expressed in a wide range of tissue types; why does it only lead to retinoblastomas and osteosarcomas?
- part of answer (known to date): other tissues require more mutations; Rb deletion is not enough to induce tumor growth
Loss-of-heterozygosity events can be used to find TSGs#
- restriction fragment length polymorphism. a polymorphic sequence that permits or disallows cleavage by a restriction enzyme
- tracking anonymous DNA sequences with known levels of heterozygosity in population can be used to find LOH events, which can be linked to nearby TSGs
Promoter methylation represents an important mechanism for inactivating TSGs#
- cytosine in CpG can be methylated (denoted meCpG)
- methylation of CpGs and the accessibility of promoter DNA are important methods cancer uses to shut down TSGs
- methylation patterns in cancer (compared to normal cells)
- in early stages of tumor development
- methylation is decreased (global hypomethylation)
- some CpG islands are hypermethylated
- 60-70% genes have CpG islands affiliated with their promoters
- in early stages of tumor development
- methylation can also undergo LOH patterns
TSGs and their encoded proteins function in diverse ways#
- tumor viruses encode proteins that disrupt function of pRB and p53 (two common TSGs), which helps explain their dominant phenotype
The NF1 protein acts as a negative regulator of Ras signaling#
- plexiform neurofibromas neurofibrosarcomas
- increased risk of gliomas, pheochromocytomas, and myelogenous leukemias
- Nf1 has high sequence homology with Ras-GAPs (GTPase activating proteins)
- Ras-GAPs negatively regulate Ras proteins by activating the GTPase activity of Ras, which stops Ras' function
- evidence indicates that
- Nf1 LOH primarily targets undifferentiated neural crest-derived cells related to Schwann cell precursors
- once such cells lose all Nf1 function, they initiate development of other tumor growths by inducing coproliferation via paracrine signaling
Cre-Lox uses flanking LoxP sites and the Cre recombinase to knockout a gene conditionally (e.g. tissue-specific knockout). This can be done because the Cre recombinase is only expressed under a tissue-specific promoter.
The alleles that are modified to have flanking LoxP sites are called floxed alleles. They are often denoted , e.g. . There is no specific allele type (e.g. ) since the allele is controllable by the experimenter.
- recruitment of mast cells to incipient neurofibromas is critical to formation of tumors
- when surrounding mast cells have genotype, neurofibromas are likely to form. If (wild-type), neurofibromas are much less likely to form.
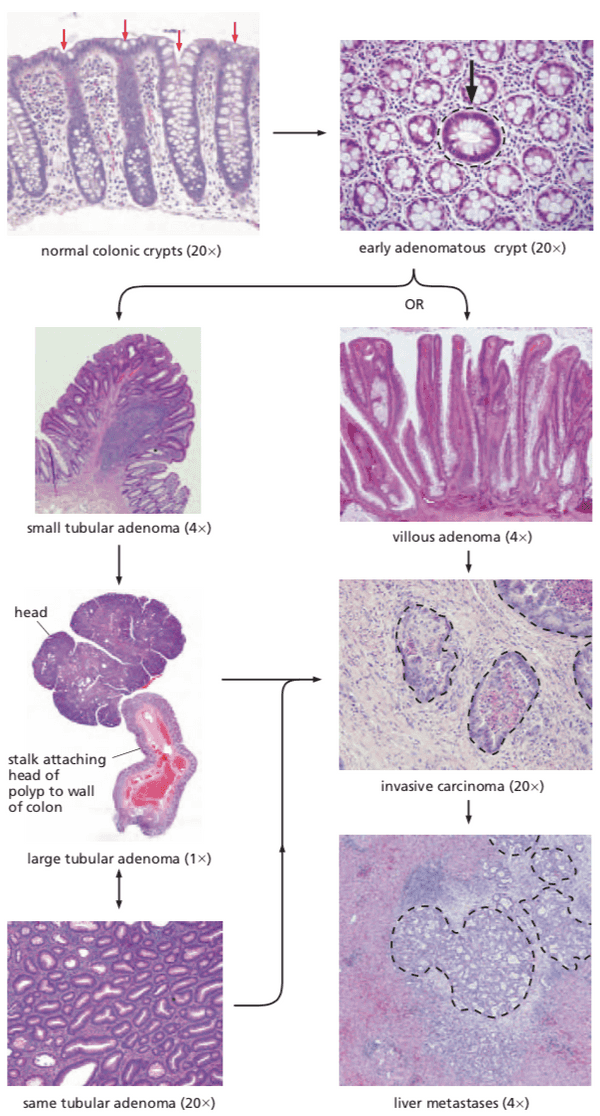
APC facilitates egress of cells from colonic crypts#
- >95% colon cancers appear to be sporadic
- one type is hereditary nonpolyposis colon cancer (=HNPCC, Lynch syndrome)
- another type is adenomatous polyposis coli (=APC, familial adenomatous polyposis, FAP)
- caused by the Apc gene
- outmigration and death in colon takes only 3 to 4 days
- the -catenin protein governs most of the outmigration behavior
- high Wnt concentration is what keeps cells in stem cell phenotype
- Wnt binding (to cell surface receptors) prevents -catenin destruction
- -catenin associated with DNA-binding proteins called Tcf/Lef
- For enterocytes, Tcf/Lef activate expression of genes that program the stem cell phenotype
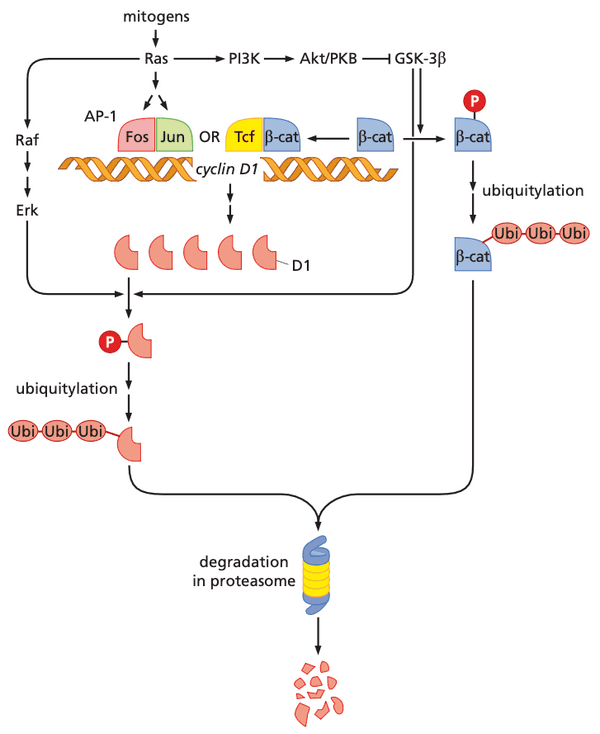
- APC protein negatively controls the level of -catenin in the cytosol (APC is inhibited by Wnt signaling)
- APC forms complex (with scaffold protein axin) bringing together GSK-3β with -catenin
- -catenin gets phosphorylated at four N-terminal residues, leading to degradation by ubiquitin-proteasome pathway
- mutations in APC prevent it from binding to -catenin and axin
- APC forms complex (with scaffold protein axin) bringing together GSK-3β with -catenin
- surprisingly, when APC functionality is recovered, tumors regress
KEAP1 regulates cellular response to oxidative stress#
- TSGs can also be linked to processes related to disease progression
- KEAP1 is a sensor for chemical and environmental stresses
- KEAP1 normally targets NRF2 for ubiquitylation in the cytoplasm; NRF2 has lifetime of 20 minutes
- ROS react with cysteines on KEAP1 KEAP1 release NRF2 NRF2 translocates to the nucleus, joins with MAF transcription factor family activate genes involved in cellular detoxification
- when ROS are cleared, cysteine residues are restored and KEAP1 continues to sequester NRF2
- some cancers disable KEAP1 so that NRF2 is constitutively activating ROS detoxification genes
- cancer is more resistant to radio- and chemotherapy
- mutations on NRF2 that prevent NRF2 from binding to KEAP1 also enable constitutive ROS detoxification
- inactivating NRF2 can make cancer formation easier (since ROS can cause mutations) but slow down cancer progression since cancers are generally in high oxidative stress environments (and need NRF2 detoxification to continue to survive)
Not all familial cancers can be explained by inheritance of mutant TSGs#
- two classes of familial cancer genes
- TSGs that directly control biology of cells (proliferation, differentiation, and death); also called gatekeepers
- genome maintenance genes that prevent mutations; called caretakers
- oncogenes are usually not transmitted through the germ line
- reason: oncogenes are dominant; usually embryonically lethal
- if LOH is 1 in , why don't people have millions of tumors?
- reason: usually mutant gene is necessary but not sufficient for tumor growth; tumorigenesis is a multi-step process
3L. Oncogenes; Introduction to Tumor Suppressor Genes#
Oncogenes#
- phenotypes are dominant
- at least 200 proto-oncogenes in the human genome
- "activating" mutations (="gain of function" mutations) convert proto-oncogene into an oncogene
- many are involved in signaling and cell cycle
- increase in function can stem from
- increased expression
- gene amplification (multiple copies of gene; double minutes or HSR)
- mutation causing weak promoter strong promoter
- introduction of a new promoter by translocation
- e.g. IgH promoter + myc
- increased protein activity
- point mutations
- deletion in growth-factor receptor
- fusion proteins
- e.g. Philadephlia chromosome (Bcr-Abl protein)
- chromosomal inversion
- e.g. EML4-ALK lung cancer; ALK kinase is no longer regulated
- WHI-P154 small molecule has inhibitory effects on ALK
- e.g. EML4-ALK lung cancer; ALK kinase is no longer regulated
- increased expression
- how can oncogenes transform when multiple mutations are needed?
- 3T3 cells are immortalized, not "normal"
- normal cells, when transformed, undergo one of the following
- no effect
- cell cycle arrest
- apoptosis
- normal cells need multiple oncogenes transfected before undergoing transformation
Tumor suppressor genes (TSGs)#
- phenotypes are recessive
- gene conversion is more likely to occur during DNA repair
- other mechanisms of TSG inactivation
- promoter methylation
- 1 mutated allele, 1 silenced allele
- 2 silenced alleles
- generation of "dominant negative alleles"
- single mutations that can eliminate functionality of other alleles, e.g. if protein works as an oligomer
- e.g. oligomeric functioning protein p53
- inhibition by other proteins
- MDM2 (ubiquitin ligase) inhibits p53
- MDM2 amplified in cancer eliminates p53
- MDM2 (ubiquitin ligase) inhibits p53
- inhibition by viral proteins
- e.g. HPV E7 inhibits pRB; HPV E6 inhibits p53 to prevent cell cycle arrest
- promoter methylation
4. Familial cancer syndromes; genetic and non-genetic risk factors#
- Readings
- pp. 386-387
- pp. 439-442
Cell death and the American way of life#
- obesity connected with increased risk of cancer
- in hormone-responsive tissue, potential mechanism: obesity increased hormone production proliferation of epithelial cells in the endometrium
- in non-hormone-responsive tissue, potential mechanism: hyperinsulinemia increased synthesis of IGF-1, reduced production of IGF-1 antagonists in the liver (IGF-binding protein 1, IGFBP-1) IGF-1 binding activates Akt/PKB, providing strong anti-apoptotic signals
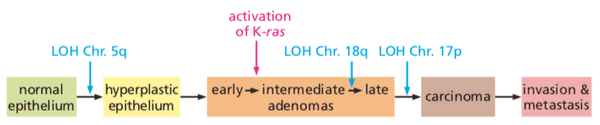
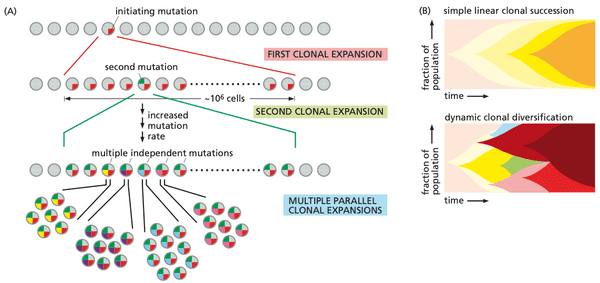
@weinberg2e/11.1 Most human cancers develop over many decades of time#
- tumor progression. the process in which normal cells evolve into cells with increasingly neoplastic phenotypes
- leading question: How many different sequential changes are actually required in cells and tissues in order to create a human cancer?
- the late onset of most cancers means that curing all cancers will have little effect on expected life span
- risk of death from cancer is to (where is age).
- means that generally 6 to 7 critical events are needed for tumor to form.
5. Shaping the cancer genome @weinberg2e/12#
- Readings
- pp. 511-573
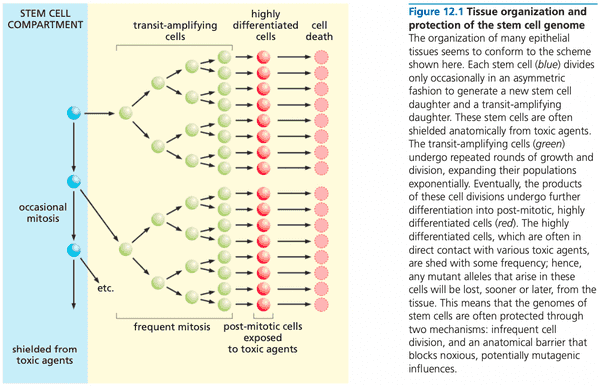
Tissues are organized to minimize the progressive accumulation of mutations#
- the stem cell compartment is shielded anatomically from toxic agents
- inevitably, some stem cells are lost and must be replaced via symmetric division, or transit-amplifying cell must return back to a stem cell
- genome might not be perfectly maintained
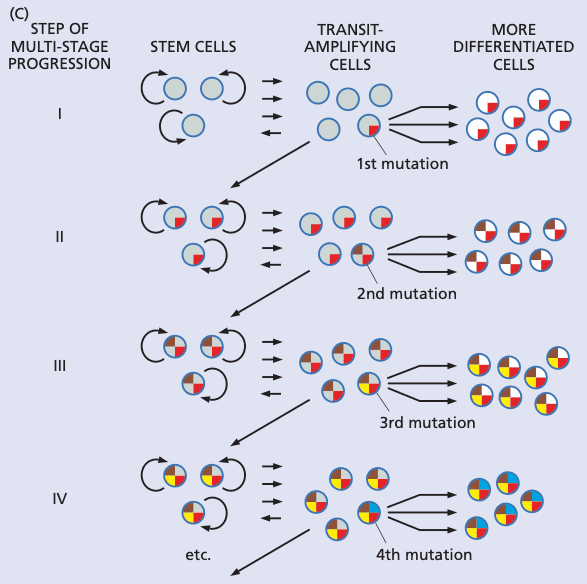
Stem cells may or may not be the targets of the mutagenesis that leads to cancer#
- treating mice with carcinogen, then treating them with 5-FU (kills actively dividing cells) didn't stop progression of cancer cancer initiator is a cell type that only divides occasionally
- blocked differentiation is a common theme in the development of blood cancers
- we don't know the precise identities of stem cells targets of transformation
- could be puripotent hematopoietic stem cell
- but could also be an early committed progenitor, and mutation is stored long-term via de-differentiation back to a stem cell
- this is a hypothesis; it is not known
- can happen if stem cells are killed and need to be resupplied via de-differentiation
- we don't know the precise identities of stem cells targets of transformation
Apoptosis, drug pumps, and DNA replication mechanisms offer tissues a way to minimize the accumulation of mutant stem cells#
- stem cells in mouse crypts, in response to DNA damage, are primed to activate apoptosis rather than risk replicating errors from repair
- it is possible some stem cells have stronger DNA repair machinery; this is hypothesized since some cancer stem cells are resistant to apoptosis and have enhanced DNA repair mechanisms
- stem cells have enhanced drug pumps
- stem cells efficiently pump out certain fluorescent dye molecules
- due to plasma membrane protein called Mdr1 (multi-drug resistance 1)
Cell genomes are threatened by errors made during DNA replication#
- assaults on DNA
- errors from DNA polymerases, including incorporation of nucleotide analogs
- spontaneous chemical changes modifying bases
- mutagenic agents (both endogenous and exogenous)
- pol- has proofreading function (3>5 exonuclease activity)
- mutation rate 1 in
- mismatch repair enzymes (=MMR) follow DNA polymerases
- especially important in regions with repeated sequences
- DNA slippage may cause DNA polymerase to create repeats
- single strand nicks indicate which strand is newly synthesized
- miss 1 mutation out of every 100; two rounds for rate of 1 out of
- double-strand DNA breaks (DSBs) can also happen during replication
- as many as 10 ds DNA breaks occur per cell genome each time a cell passes through S phase
- ds breaks occur near replication forks, likely because ssDNA unwound but not-yet-replicated is susceptible to inadvertent breakage
- as many as 10 ds DNA breaks occur per cell genome each time a cell passes through S phase
Cell genomes are under constant attack from endogenous biochemical processes#
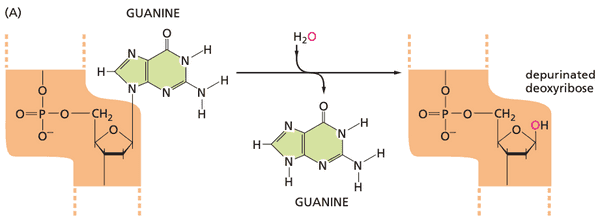
- hydrogen and hydroxyl ions in water can cause accidental DNA damage, such as depurination, in which the bond linking a purine base to the deoxyribose breaks
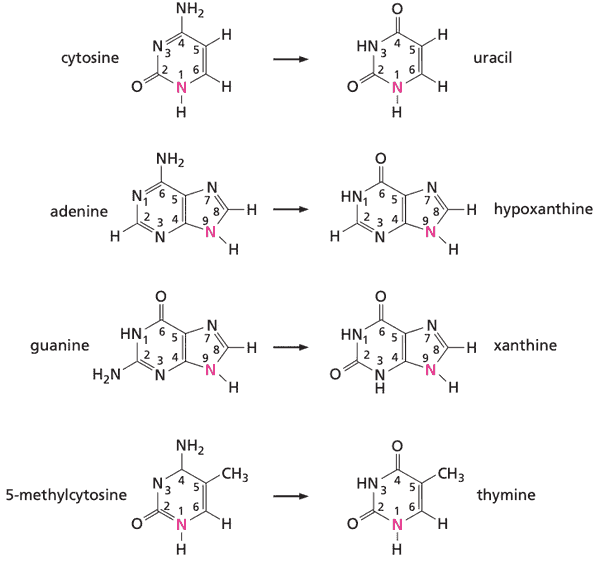
- deamination of 5-methylcytosine can evade detection because its resultant base is thymine, a normal base (i.e. can cause C>T point mutations)
- ROS generated from reduction of oxygen to water can also cause damage
- estimated 1-2% oxygen molecules consumed by mitochondria end up as ROS
- increased production in inflammation
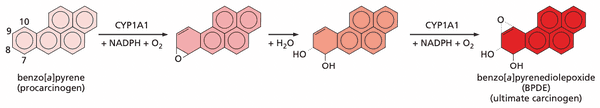
Cell genomes are under occasional attack from exogenous mutagens and their metabolites#
- x-rays form ROS from water
- UV photons often cause pyrimidine dimers, covalent bonds between two adjacent pyrimidines on the same strand of DNA
- cytosines become prone to deamination, leading to a CC TT transition
- electrophilic chemical species ingested are mutagenic
- example: alkylating agents
- BPDE
- acetaldehyde
- aflatoxin B1
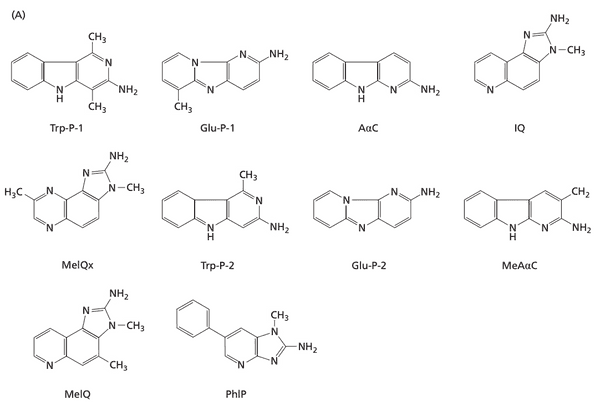
- heterocyclic amines (HCA)
- array of cytochrome P450 enzymes (CYPs) designed to oxidize PAHs to make easier to secrete
- can inadvertently turn procarcinogens (hydrocarbons are generally inert) into active carcinogens
- ethanol's mutagenicity stems from acetaldehyde, which can react with deoxyguanosine, forming several DNA adducts
- pediatric cancers have ~1 point mutation per Mb
- smoking-related cancers and melanomas have ~10 point mutations per Mb.
- heterocyclic amines (HCAs) from eating meat cooked at high temperatures
- PhIP. 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
Cells deploy a variety of defenses to protect DNA molecules from attack by mutagens#
- most effective way for a cell to defend its genome: physical shielding
- skin as barrier for internal cells
- melanin as barrier for skin
- chemical interventions
- ROS scavengers
- superoxide dismutase
- catalase
- vitamin C
- -tocopherol (vitamin E)
- bilirubin
- urate
- 90% of prostate carcinomas show the loss of expression of GST-
- traceable to the methylation of the GSTP1 promoter
- ROS scavengers
Repair enzymes fix DNA that has been altered by mutagens#
- more than 160 proteins responsible for repairing DNA
- technique 1: enzymes catalyze reverse reaction that resulted in altered base
- e.g. -methylguanine-DNA methyltransferase (MGMT, =DNA alkyltransferase)
- removes methyl and ethyl adducts at position of guanine
- can blunt the cytotoxic effects of alkylating chemotherapy drugs!
- removes methyl and ethyl adducts at position of guanine
- e.g. -methylguanine-DNA methyltransferase (MGMT, =DNA alkyltransferase)
- technique 2: base excision repair (BER)
- tends to be used for lesions from endogenous sources
- glycosylases cleave recognize and cleave abnormal bases
- e.g. uracil DNA glycosylases
- e.g. T:G DNA glycosylases
- AP endonuclease (=APE, apurinic/apyridinic endonuclease) with AP lyase removes base-free sugars (APE cleaves 5' end; APL cleaves 3' end)
- technique 3: nucleotide excision repair (NER)
- generally repairs lesions from exogenous sources
- focuses on bulky, helix-distroying alterations
- excises ~29 nt: 24 nt on 5' side, 5 nt on 3' side
- two types of repair
- transcription-coupled repair (TCR). performed on DNA template strands that are actively transcribed
- global genomic repair (GGR). performed on DNA non-template strands as well as non-coding regions of DNA
- p53 activates expression of GGR proteins
- technique 4: bypass synthesis with error-prone polymerases
Inherited defects in mucleotide-excision repair, base-excision repair, and mismatch repair lead to specific cancer susceptibility syndromes#
- xeroderma pigmentosum is a result of defective NER
- 7/8 XP-associated genes: XP{A,B,C,D,E,F,G} encode components of the NER complex
- XPV encodes error-prone DNA polymerase pol- used when regular DNA polymerases (e.g. pol-) are unable to copy over unrepaired DNA lesions (e.g. pyrimidine dimers).
- hereditary non-polyposis colon cancer (=HNPCC). another cancer susceptibility syndrome caused by inherited defects in DNA repair
- mismatch repair genes are defective
- 85-90% cases: MSH2 and MLH1
- 15% cases: MSH6 and PMS2
- leads to high mutation rates in genes with microsatellite repeats
- e.g. type II TGF- receptor (TGF-RII)
- ; inhibition by TGF growth factor no longer occurs
- e.g. type II TGF- receptor (TGF-RII)
A variety of other DNA repair defects confer increased cancer susceptibility through poorly understood mechanisms#
- BRCA1 and BRCA2
- susceptibility to breast and ovarian carcinomas
- evidence for participating in genomic maintenance is guilt-by-association
- found in complexes with RAD50/Mre11 and RAD51 proteins, which are homologs of proteins in yeast involved in repairing DNA breaks caused by x-rays
- BRCA2 partial loss of function
- high rates of illegitimate recombination (between chromosomal arms that are nonhomologous)
- NHEJ takes over if HDR fails
- NHEJ is actually plays a major role in VDJ recombination
- also enables class switching between immunoglobulin types
- deregulation of centrosome number
- high rates of illegitimate recombination (between chromosomal arms that are nonhomologous)
- BRCA1/BRCA2 breast cancers that initially respond to cisplatin therapy develop resistance by developing back-mutations that restore the function of the BRCA1/2.
- BRCA1 likely a scaffold for DNA repair complexes
- BRCA1 loss H2A not ubiquitylated H2A no longer heterochromatin, variety of repeat sequences expressed genetic destabilization
The karyotype of cancer cells is often changed through alterations in chromosome structure#
- aneuploid. deviation in chromosome number
- chromosomal aberration. changes in chromosome structure of individual chromosomes
- molecular mechanisms that lead to chromosomal aberrations not known
- possibility: breakage-fusion-breakage (BFB) cycles
- used to be believe that translocations were only in blood tumors
- 2005, discovered translocation TMPRSS2/ERG, present in 50% of localized prostate carcinomas
- genome sequencing led to discovery of chromothripsis (chromosome shattering)
- mechanism unknown
The karyotype of cancer cells is often changed through alterations in chromosome number#
- chromosomal instability (CIN). phenomenon where a population of (usually cancer) cells have a large distribution of chromosome copy numbers
- both CIN and microsatellite instability (MIN) provide mutability for cancer tumor growth
- changes in chromosome number are usually consequences of mis-segregation of chromosomes during mitosis (nondisjunction)
- can be caused by failure of quality control ensuring chromatids are attached to their spindle fibers
- another source is kinetochores (the disk-shaped center located on the chromosomes that attaches to spindle fibers) containing too many spindle fibers (normal is 20-25 microtubules)
- leads to merotely where a kinetochore is attached to spindle fibers on both sides of the dividing cell, resulting in the loss of the chromosome
- spindle assembly checkpoint (SAC) fails to check correct attachment of kinetochores to chromatids
- leads to merotely where a kinetochore is attached to spindle fibers on both sides of the dividing cell, resulting in the loss of the chromosome
- can also be caused by incorrect spindle assembly, such as supernumerary centrosomes at interphase
- CIN continues even after tumor progression has completed (unlike breakage-fusion-bridge cycles, which happen during progression and then stop)
- molecular defaults leading to CIN
- duplication of centrosomes occurs at G1/S transition
- HPV E7 protein can destabilize centrosome number via pRb loss-of-function
- HPV E6 protein allows cell to tolerate centrosome abnormalities by disrupting p53
- duplication of centrosomes occurs at G1/S transition
- common cancers can be caused by inherited defects in caretaker genes
- healthy cells taken from cancer patients (but not cancer cells) are more vulnerable to ionizing radiation than healthy cells taken from healthy control patients
Thought questions#
- What types of evidence suggest that karyotypic alterations of cell genomes are not absolutely essential for neoplastic transformation?
- When calculating the rates of mutation required in order for multi-step tumor progression to reach completion, what parameters must one know in order for such a calculation to accurately describe the actual biological process?
- How does our understanding of defective DNA repair processes in tumor cells make possible the development of new anti-cancer therapeutic strategies?
- In which ways do the defectiveness of p53 function and resulting defects in apoptosis and DNA repair facilitate the forward march of tumor progression?
- What types of tumor promotion, as described in Chapter 11, favor the genetic evolution of premalignant cell clones?
- What evidence implicates mutagenic chemicals originating outside the body in the pathogenesis of human cancers? How can one gauge their contribution to human carcinogenesis compared with that of mutagens and mutagenic processes of endogenous origin?
- How do defects in various cell cycle checkpoints allow for accelerated rates of the accumulation of mutations?
- How do the biological properties of stem cells help to reduce the rate at which tissues accumulate mutant genes?
- How does the existence of cancer stem cells affect the calculations of the rate at which mutations must be accumulated in order to allow multi-step tumor progression to advance?
- How does the genetic heterogeneity in the human gene pool affect the functioning of various types of biological defenses that have been erected to prevent the accumulation of mutant alleles in human somatic cells?
6. Characterizing the cancer genome#
Exploring the genomes of cancer cells: progress and promise#
- first generation DNA sequencing: Sanger sequencing
- second generation DNA sequencing: pyrosequencing (454), Illumina
- third generation DNA sequencing: nanopore sequencing
- usually 1,000 to 10,000 somatic substitutions in most adult cancers
- lung cancer and melanomas can have upwards of 100,000
Mutational signatures: the patterns of somatic mutations hidden in cancer genomes#
- matching mutational patterns induced by carcinogens and the mutations found in particular cancers can reveal potential causes of cancer (bridging molecular cancer genetics and epidemiology)
- mutational patterns = mutational spectra
- devised algorithm to decompose mutational spectra into multiple mutation processes (which can be correlated to a causative agent)
7. Functional genomics#
8. Epigenetic and cancer development#
- Readings
- Weinberg, pp. 21-24, 249-254
Histone modification and transcription factors control gene expression#
- transcription factors bind to enhancers and silencers
- activators transcription factors that bind to enhancers
- repressors transcription factors that bind to silencers
- RNA pol II generally synthesizes both strands, and then is paused in transcriptional pausing before other signals either allow or prevent it from resuming elongation
Promoter methylation represents an important mechanism for inactivating tumor suppressor genes#
- mammalian cells can have methylation on the cytosine in CpG sites (denoted MeCpG)
- often causes repression of transcription
- cytosine is the base that is methylated
- mechanisms not entirely known
- one mechanism: protein complex can bind MeCpGs and second subunit functions as histone
- de novo methylation. where methylation of histones can lead to methylation of CpGs
- cancer is associated with two patterns of methylation
- global _hypo_methylation
- _hyper_methylation at CpG islands
- CpG islands are often affiliated with gene promoters (~70% genes have promoters with CpG islands)
- some cancer cells in tumors can have ~5% genes hypermethylated
- may be due overexpression of DNA methyltransferase 3B (DNMT3B)
- example of physiological consequence of methylation
- RAR2 gene encodes a retinoic acid receptor that arrests the cell cycle in the presence of retinoic acid. methylation of this promoter prevents cells from responding to retinoic acid.
9. Signal transduction (I)#
- Readings
- Weinberg, The Biology of Cancer 2e, pp. 131-174
@weinberg2e/5.1 Normal metazoan cells control each other's lives#
- lots of communication between cells in a tissue is via growth factors
- growth factor in serum that promotes clotting: platelet-derived growth factor (PDGF)
- PDGF attracts fibroblasts and then stimulates their proliferation
- many oncogenes encode growth factor receptors; mutations in them trick cells into believing they have encountered large concentrations of growth factor
The Src protein functions as a tyrosine kinase#
- used antibodies to show that Src was kinase
- independent of kinase function, Src is also a phosphoprotein (carries phosphate groups attached covalently to one or more of its amino acid side chains).
- tyrosine phosphorylation is used largely by mitogenic signaling pathways
- serine/threonine phosphorylation are used by other kinases (non-mitogenic signaling pathways)
- the v-src lead to significant increase in concentration of phosphotyrosines
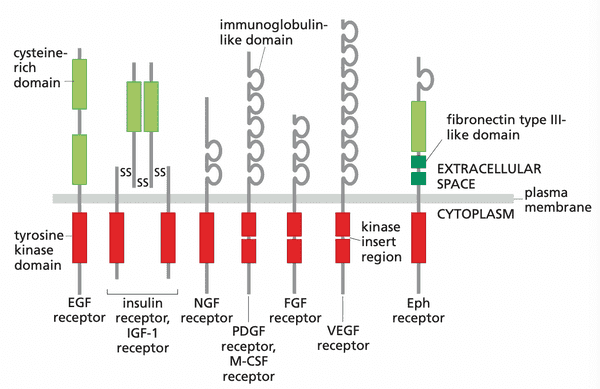
The EGF receptor functions as a tyrosine kinase#
- epidermal growth factor (EGF) was one of the first growth factors discovered (nerve-derived growth factor, NGF, was the first)
- EGFR found to be a receptor tyrosine kinase (RTK)
- ectodomain. the domain that projects into the extracellular space
- intracellular domain of EGFR acts as Src-like kinase and signals cell proliferation
- changes that can be activated by growth factors
- cell growth and division
- cell shape
- cell survival
- cell motility
- RTKs likely evolved right before metazoan life since they are common to all metazoa
An altered growth factor receptor can function as an oncoprotein#
- erbB oncogene was truncated version of EGFR transmit cell proliferation signals even in absence of growth factors
A growth factor gene can become an oncogene: the case of sis#
- v-sis is very similar to the B chain of PDGF (platelet-derived growth factor)
- Friend leukemia virus, env gene is actually encodes a EPO (erythropoietin) growth factor mimic
- a cell that produces a growth factor it responds to leads to autocrine signaling of proliferation
- common growth factors with autocrine signaling
- TGF-
- stem cell factor (SCF)
- insulin-like growth factor (IGF-1)
- tumors with autocrine signaling are more likely to have in vivo results that match in vitro experiments, likely because they create their own growth factor environment
Transphosphorylation underlies the operations of receptor tyrosine kinases#
- RTKs form dimers and transphosphorylate
- overexpression of receptors can lead to spontaneous dimerization, or dimerization that sensitizes the complex to ligand binding
- point mutations in the transmembrane domain can increase the affinity of RTKs for each other
- RTKs can be mutated to have constitutively activated kinases
- fusion proteins whose newly added fusion component has high affinity for dimerization can increase likelihood of RTK dimerization
- kit is receptor for SCF (stem cell factor)
- at least 59/20000 genes encode RTK-like proteins
- receptor genes can be transmitted in the human germ line
- mutants are often a recessive allele
- single dominant allele can sustain development because loss of the dominant allele is infrequent
- some dominant alleles cause developmental problems but not death, but result in death before adulthood
- these are generally de novo mutations
Yet other types of receptors enable mammalian cells to communicate with their environment#
Cytokine receptors#
- examples include
- EPO receptors
- TPO (thrombopoietin) receptors
- controls the development of megakaryocytes, the precursor of platelets
- interferon receptors
- interleukin receptors
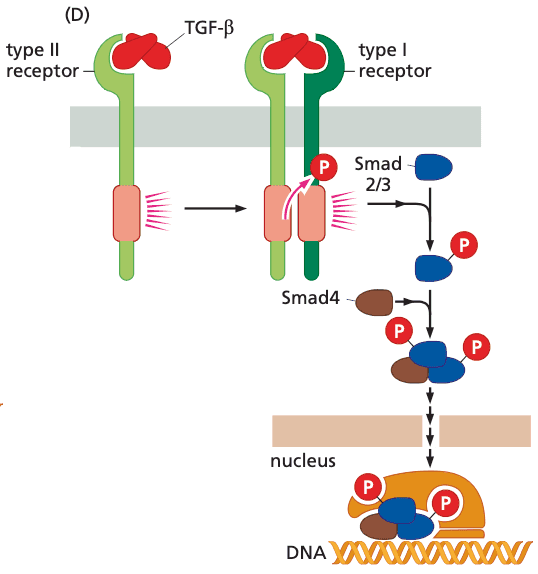
TGF- receptor#
- structure
- invariably function as heterodimers
- have serine/threonine kinase domains rather than tyrosine
- ser/thr kinase is constitutively active
- function of TGF-
- suppress proliferation of normal epithelial cells
- promote acquisition of invasive properties by already-transformed cells
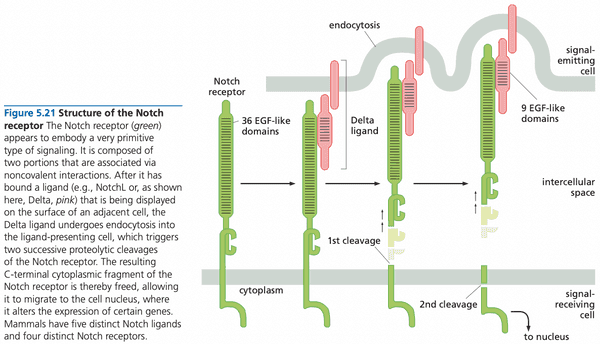
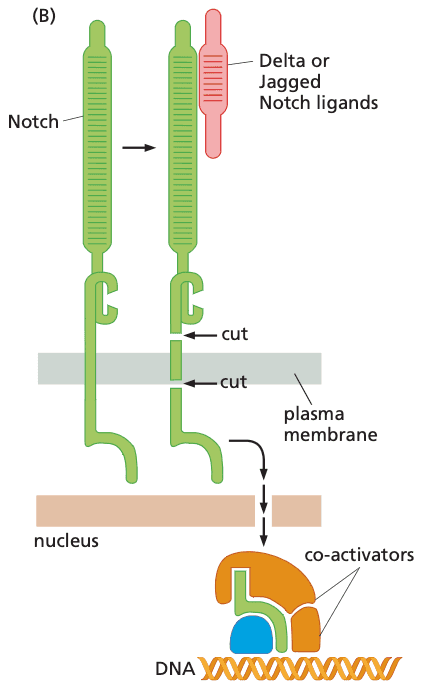
Notch#
- Notch is the receptor
- Notch ligands
- examples include
- NotchL
- Delta
- Jagged
- Notch ligands are immobilized cell surface proteins cell-cell physical interactions; juxtacrine signaling
- examples include
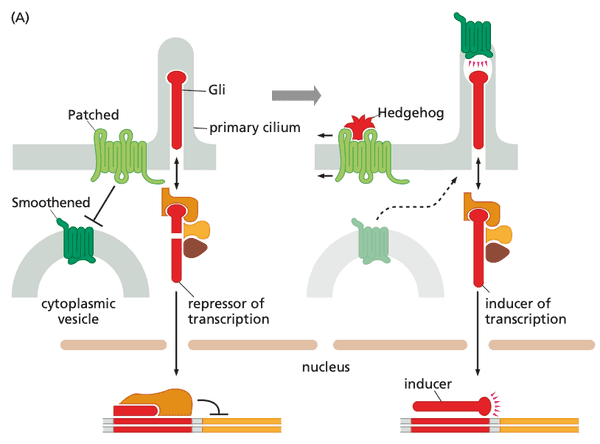
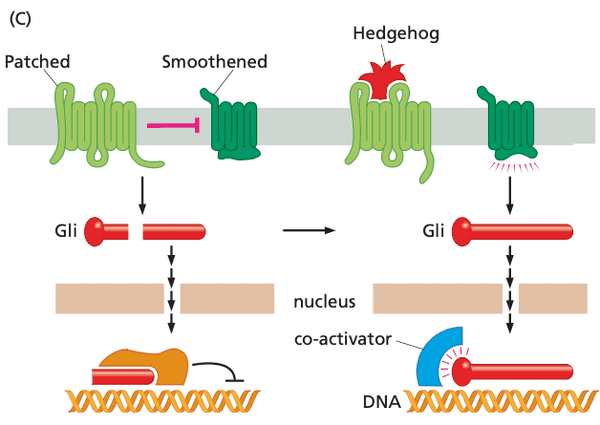
Patched-Smoothened (Ptc-Smo)#
- ligands include
- proteins of Hedgehog (Hh) class
- system description
- Patched sequesters Smoothened from entering the primary cilium
- Gli in the primary cilium thus get cleaved and enters the nucleus to repress transcription
- when Hedgehog binds to Patched, Smoothened travels to primary cilium and prevents Gli from cleavage
- Gli moves to nucleus but now functions as an inducer of transcription
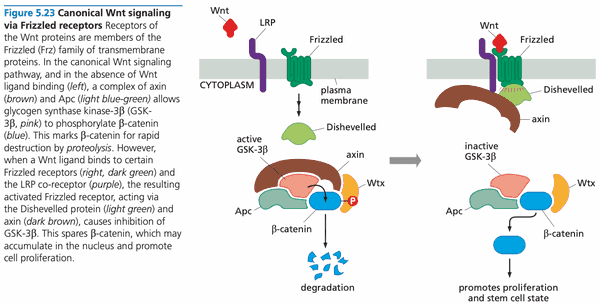
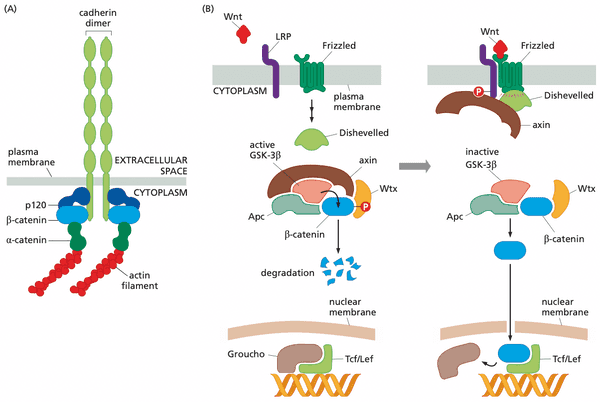
Wnt signaling system#
- Humans make at least 19 types of Wnt proteins (=Wnt growth factors)
- Wnt proteins are tethered tightly to the ECM and (through lipid tail) to cell membranes not freely diffusible like other growth factors
- Wnt proteins activate Frizzled (Frz) family of receptors
- "canonical Wnt signaling"
- no Wnt
- glycogen synthase kinase-3 (GSK-3) phosphorylates -catenin, marking it for destruction
- GSK-3 requires complex of axin, Wtx (Wilms tumor protein), and Apc to function
- with Wnt
- Frz binds Dishevelled and axin, preventing formation of complex needed for GSK-3 to function; -catenin not degraded
- -catenin promotes cell proliferation
- no Wnt
- non-canonical Wnt signaling
- non-canonical Frz receptors activate G-proteins (Frz receptors are GPCRs)
Nuclear receptors sense the presence of low-molecular-weight lipophilic ligands#
- low-molecular weight lipophilic ligands: steroids, retinoids, vitamin D
- esterogen, progesterone, and androgen receptors
- play key roles in development of breast, ovarian, and prostate carcinomas.
- structure of nuclear receptor molecules
- DNA-binding domain
- recognizes hormone response elements (HREs) in the DNA
- pair of hexanucleotides separated by a variable number of spacer sequences
- hinge region
- conserved ligand-binding domain
- DNA-binding domain
- tamoxifen (a selective estrogen receptor modulator) works by binding nuclear receptor and causing conformation change so that the receptor cannot act as a co-activator.
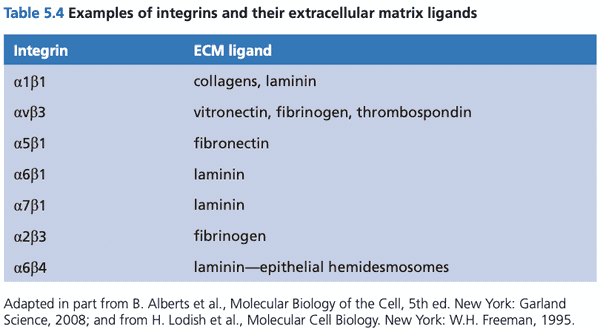
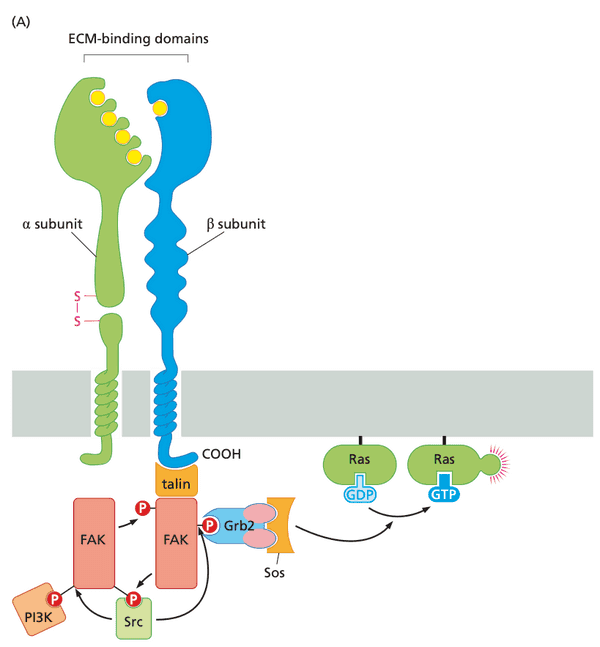
Integrin receptors sense association between the cell and the extracellular matrix#
- ECM consists of collagens, laminins, proteoglycans, and fibronectin.
- cells grown in tissue flasks secrete an ECM, then bind to the ECM.
- sensing of collagen in ECM is accomplished by discoidin domain receptors (DDR-RTKs) (the receptors are receptor tyrosine kinases)
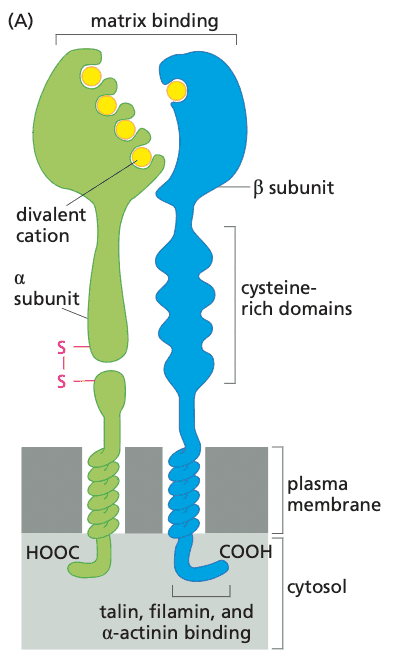
- most other sensing of ECM components is accomplished through integrins
- structure of integrins
- heterodimers consisting of and subunits
- at least 18 subunits and 8 subunits discovered
- at least 24 distinct heterodimers
- RGD receptors; recognize arginine-glycine-aspartic acid tripeptide motif
- integrins cluster to form focal adhesions
- some integrins are attached to components of the cytoskeleton, e.g.
- actinin, vinculin, talin, paxillin are the linkages
- activated integrins evoke a variety of cell responses, such as cell migration, proliferation, and survival
- unique feature of integrins: signals from inside the cell can mediate binding outside of the cell
- e.g. focal adhesion kinase (FAK) is needed to transmit signals to detach from ECM; FAK knockout leads to loss of motility
The Ras protein, an apparent component of the downstream signaling cascade, functions as a G protein#
- 3 ras genes encode for 4 Ras proteins (K-ras has a second protein from alternative splicing)
- C-termini all carry covalently attached lipid tails
- Ras mutations that prevent the function of Ras-GAP lead to overly high levels of active Ras.
10. Signal transduction (II)#
- Readings
- Weinberg, The Biology of Cancer 2e, pp. 175-229
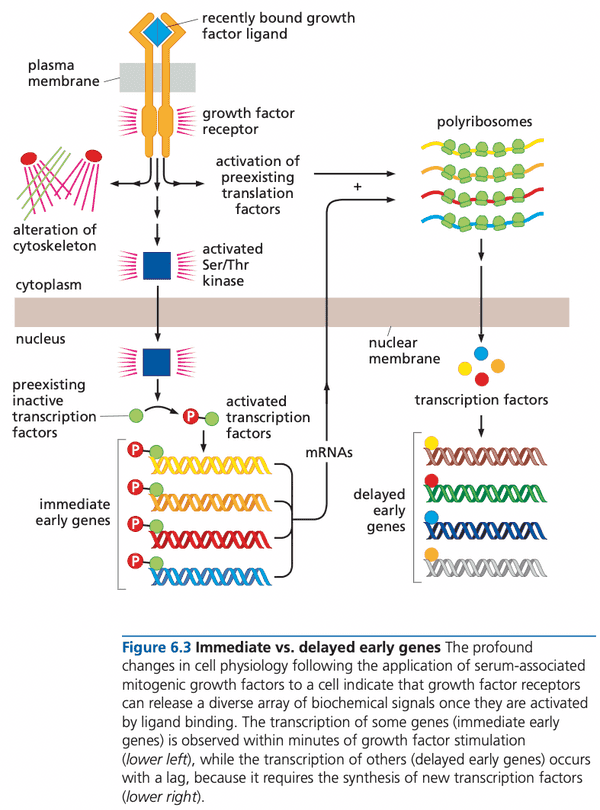
@weinberg2e/6.1 A signaling pathway reaches from the cell surface into the nucleus#
- cycloheximide. a drug that shuts down all cellular protein synthesis.
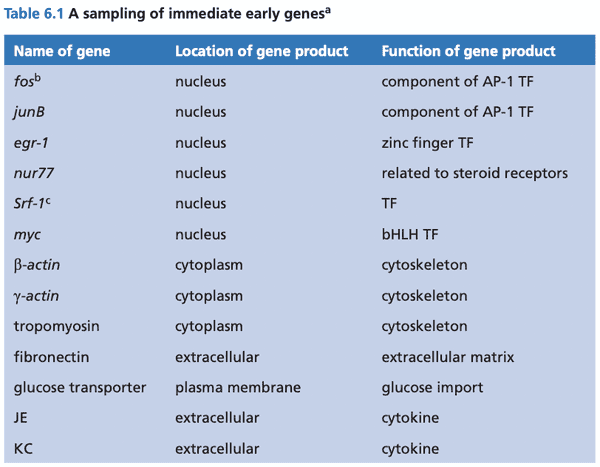
- immediate early genes (IEGs). genes whose expression increases rapidly within a half hour of growth factor stimulation
- IEG gene products help cell emerge from .
- signal induction to begin transcription of IEGs does not require new proteins to be synthesized
- Myc signals mitogen concentration by increasing its concentration
- contrast with ras and src that signal through structure changes
- other effects of growth factors
- increased rate of protein synthesis by activating proteins for ribosome binding
- reorganize actin fibers of cell's cytoskeleton
- provide survival signals to avoid inadvertent activation of apoptosis
The Ras protein stands in the middle of a complex signaling cascade#
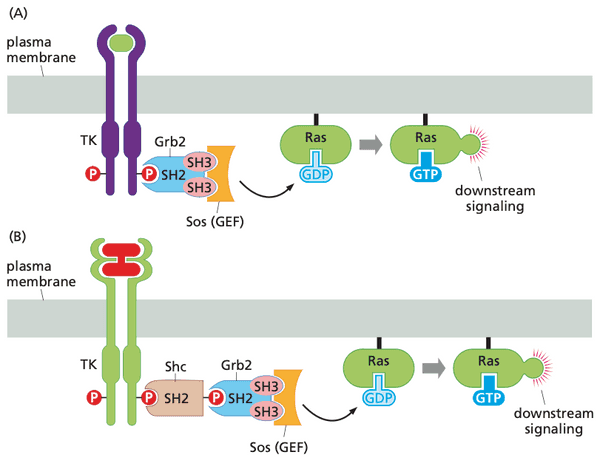
- discovered Sos through studies of Sevenless receptor for fruitfly ommatidia (light-sensing units).
- discovered that Sos was a GEF, the once-hypothetical protein that activated Ras (Ras-GEF = Sos)
Tyrosine phosphorylation controls the location and thereby the actions of many cytoplasmic signaling proteins#
- location rather than conformation controls the effects of many cytoplasmic signaling proteins
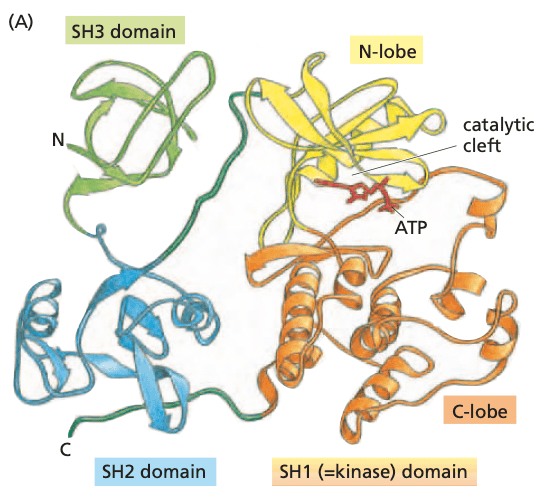
- Src protein structure
- SH1 domain. (Src homology domain 1); kinase domain with N and C lobes
- SH2 domain. binds phosphotyrosine and specific oligopeptide sequence, depending on the specific sequence for the SH2 domain
- SHP1. a phosphatase that has an SH2 domain
- cytosol. the soluble portion of the cytoplasm
- growth factor receptors trans-phosphorylate SH2-containing proteins bind to phosphorylated receptors
- some of these newly bound proteins may be phosphorylated by the receptor
- others may act as binding sites for multi-protein complexes
- SH3 binds proline-rich sequence domains in partner proteins
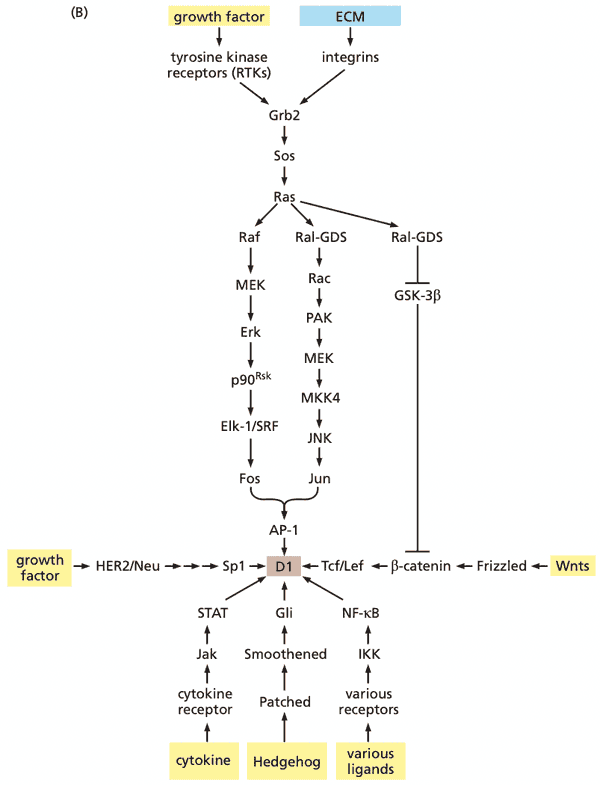
SH2 and SH3 groups explain how growth factor receptors activate Ras and acquire signaling specificity#
- Grb2 has two SH3 groups and one SH2 group; acts as a bridge protein
- SH3 domains bind Sos
- SH2 domain binds pY on growth factor receptor
- Sos is now in close proximity to activate membrane-bound Ras
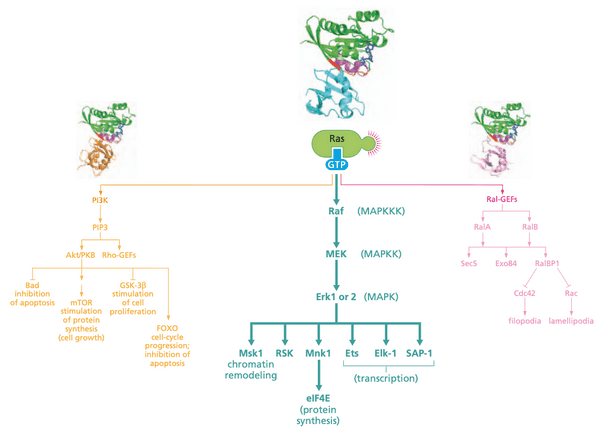
Ras-regulated signaling pathways: A cascade of kinases forms one of three important signaling pathways downstream of Ras#
- This section discusses Raf
- Ras effectors (the proteins that bind Ras and carry out the effects of activated Ras)
- Raf (serine/threonine) kinase (MAPK pathway)
- PI3K
- Ral-GEF
Raf details#
- confers anchorage-independence and loss of contact inhibition
- Raf undergoes configuration change (not translocation)
- Ras Raf (=c-Raf) MEK (=MAP2K, MAPKK, mitogen-activated protein kinase kinase) MAPK (=, extracellular signal-regulated kinase) Fos + Jun = AP-1
- MEK can phosphorylate serine, threonine, and tyrosine
- ERK phosphorylates cytoplasmic substrates but also can translocate to the nucleus
- MAPK are used to refer to the class of proteins
- cascade is held together by scaffolding proteins
- KSR1 holds Raf, MEK, and Erk together
- cell can control signal tranduction by regulating activity of scaffold proteins
- Raf pathway is responsible for majority of transforming power of Ras oncoproteins
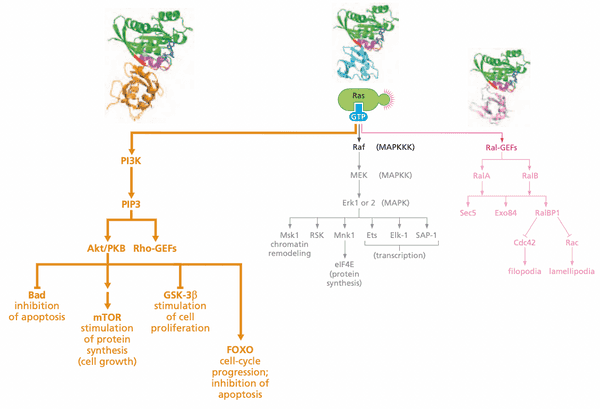
Ras-regulated signaling pathways: a second downstream pathway controls inositol lipids and the Akt/PKB kinase#
PI3K details#
- suppression of apoptosis
- PI3K has 2 subunits
- p110 catalytic subunit
- p85 regulatory subunit
- PI3K activated when p85 binds to RTK and Ras:GTP binds to p110
- RTK ligands include PDGF, NGF, IGF-1, IL-3, and integrin:ECM attachment
- some phospholipid heads have an inositol group
- phospholipase C (PLC) cleaves PI(4,5)P2 into diacylglycerol (DAG) ( PKC) and IP3 ( release of ions)
- PIP2 PI(3,4,5)P3 Akt (=PKB) apoptosis, cell proliferation, cell growth ( protein synthesis), angiogenesis
- Akt contains PH domains that bind to PIP3
-
PIP3 levels typically very low in absence of mitogens
- PTEN phosphatase removes 3' phosphate from PIP3
- PIP3 binds and activates Rho via Rho's PH domain
- affects cell motility invasiveness
- Rho proteins
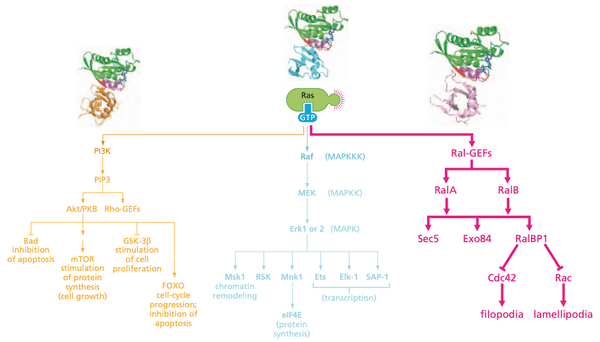
Ras-regulated signaling pathways: a third downstream pathway acts through Ral, a distance cousin of Ras#
- Ras-like proteins RalA and RalB
- Sec5, Exo84: anchorage-independent growth
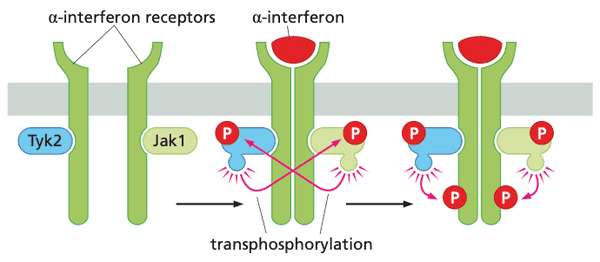
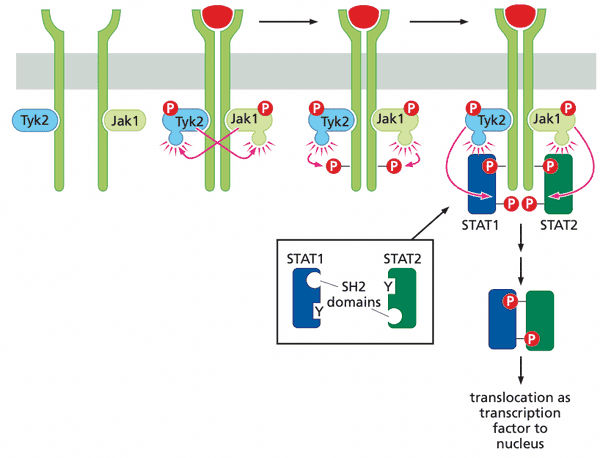
The Jak-STAT pathway allows signals to be transmitted from the plasma membrane directly to the nucleus#
- cytokine (IFN, erythropoietin=EPO, thrombopoietin=TPO) receptors complex with Janus (tyrosine) kinases (JAKs)
- Jak1, Tyk2 JAK family
- thrombopoiesis. platelet formation
- STAT = "signal transducers and activators of transcription"
- Jak-STAT pathway genes transcribed
- myc
- cyclins D2 and D3
- Bcl-XL
- stable STAT dimers (e.g. STAT3 mutants) are oncoproteins (constitutive transcription factors)
Cell adhesion receptors emit signals that converge with those released by growth factor receptors#
- focal adhesions activate focal adhesion kinase (FAK), a non-receptor tyrosine kinase
- a minimal signaling pathway would be
- ECM integrins Sos Ras Raf, PI3K, Ral-GEF
- very similar to RTK signaling!
- potential reason Ras overexpression enables anchorage-independent growth
The Wnt/-catenin pathway contributes to cell proliferation#
- at least 19 different Wnt factors in different tissues
- Wnt ligand binds Frizzled receptor to suppress activity of glycogen synthase kinase-3 (GSK-3)
- GSK-3 normally phosphorylates -catenin and tags it for destruction
- -catenin is a cytosolic protein that exists in 3 states
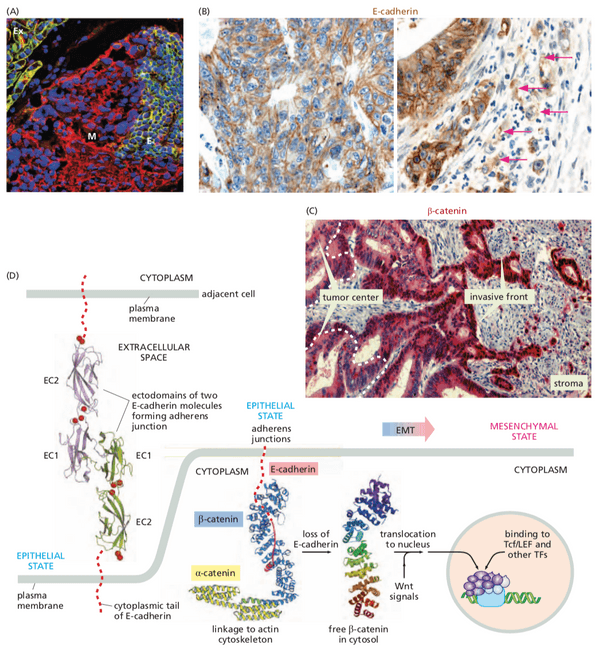
- bound to cell-cell adhesion receptors, notably E-cadherin (which forms adherens junctions)
- freely soluble in the cytosol, with a lifetime of 20 minutes
- operating in the nucleus as a component of a transcription factor (-catenin-Tcf/Lef)
- activates genes for cyclin D1 and Myc
- -catenin is a cytosolic protein that exists in 3 states
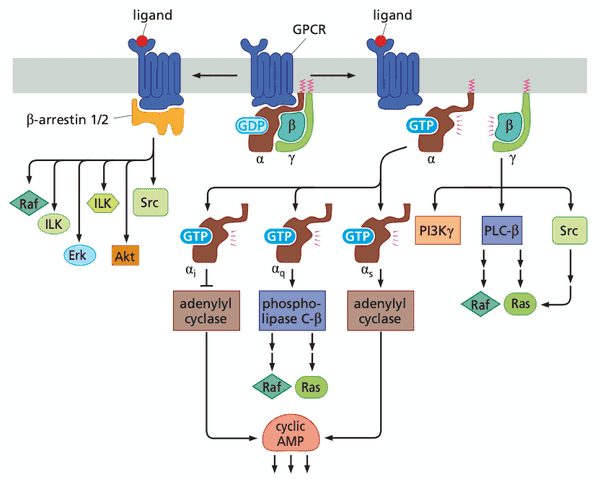
GPCRs can also drive normal and neoplastic proliferation#
- G is the subunit that binds GDP/GTP
- -arrestin downstream kinases involved in cell proliferation and survival
- arrestin in retinal cells stops activation of GPCR in response to a photon
Four additional "dual-address" signaling pathways contribute in various ways to normal and neoplastic proliferation#
- "dual address" = dispatch nuclear transcription factors that otherwise reside in the cytoplasm until signal received
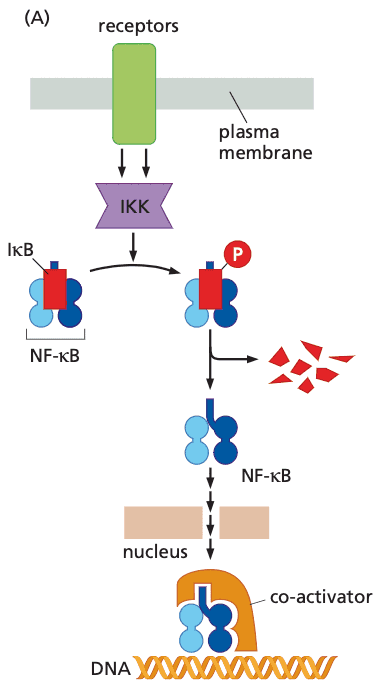
Nuclear factor-B (NFB)#
- rel NF-κB family
- NF-κB are usually sequestered in cytoplasm by IκB (inhibitor of NF-κB)
- IκB phosphorylated by IKK IκB tagged for degradation
- IKK activated by TNFα, IL-1, LPS, ROS, anti-cancer drugs, gamma radiation
- NF-κB induces expression of
- Bcl-2, IAP1, IAP-2: anti-apoptotic proteins
- myc, cyclin D1: components of cell cycle that drive proliferation
- in cancer, usually components are not mutated, but instead have constitutive expression of IKK
Notch#
- ligands: Jagged or Delta-like protein
- upon binding, receptor undergoes two irreversible proteolytic cleavages
- receptor firing occurs in direct proportion to the number of ligands encountered in the extracellular space
- mutations that delete the extracellular domain of protein lead to constitutive Notch signaling
Hedgehog#
- Patched gene inactivation Gli never cleaved constitutive activation of transcription
TGF-#
- absence of critical Smads cells can escape growth inhibitory actions of TGF-β and proliferate
- Smads bind a tetranucleotide sequence of chromosomal DNA; requires co-binding with other adjacent transcription factors to stay bound
Well-designed signaling circuits require both negative and positive feedback controls#
- negative feedback: limit function or output
- positive feedback: accelerate/amplify activation
- feedforward: maintain/stabilize a decision
11. Cell cycle @weinberg2e/8#
- Readings
- Weinberg, The Biology of Cancer 2e, pp. 275-323
Cell growth and division is coordinated by a complex array of regulators#
- G1 (first gap) phase can last between 12 to 15 hours
- S (synthesis) phase can require 6 to 8 hours
- G2 (second gap) phase ranges 3 to 5 hours
- M phase lasts around 1 hour
- other cell checkpoints include decatenation in late G2, which ensures DNA helices replicated in S phase are untangled from each other
- breakdown of control allows nascent cancer cells to acquire more mutations at a faster rate
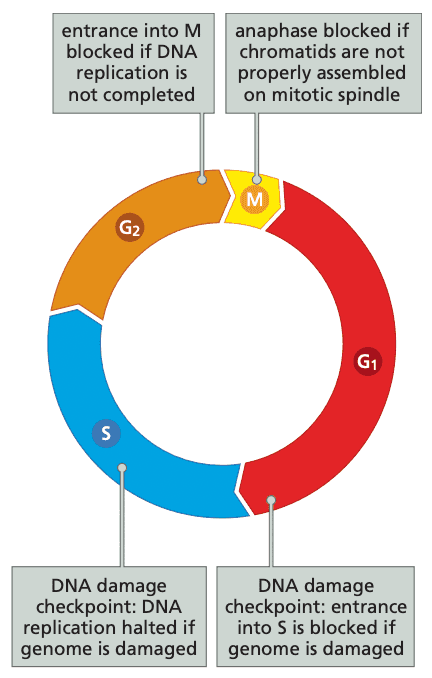
Cells make decision about growth and quiescence during a specific period in the G1 phase#
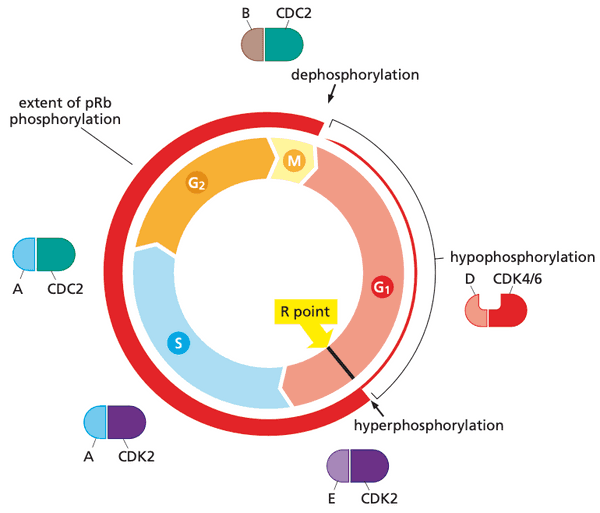
- The first 80-90% of G1 is the time period in which cells can decide to continue the cell cycle, remain in G1, or retreat into G0.
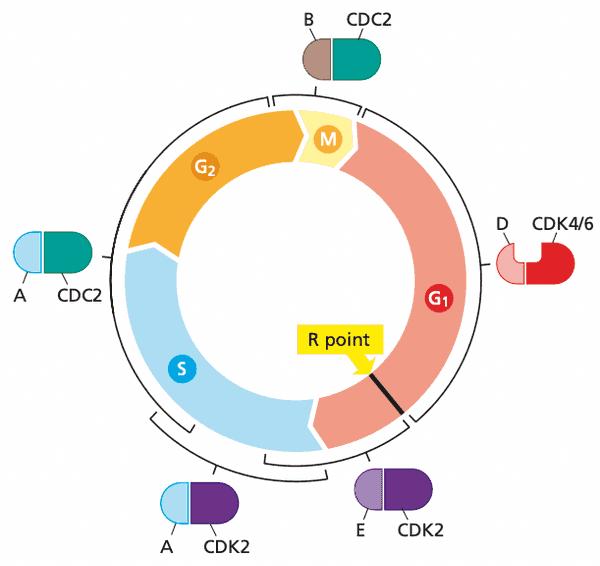
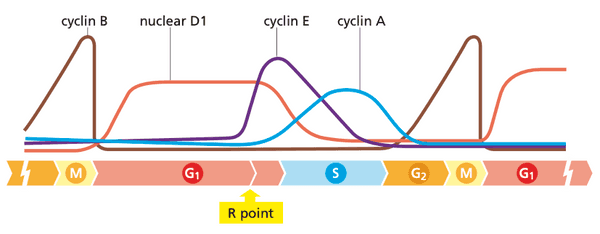
- The ending point of this decision period is called the R (restriction) point.
- The G1 S G2 M progression proceeds similar in normal and cancer cells, so the pre-R point phase is the most interesting
- cancer cells often lose their ECM attachment G1 checkpoint and are therefore anchorage-independent
- potential additional checkpoints between R point and S phase (not yet well characterized)
- checks for nutrient levels
- adequately high levels of ROS
Cyclins and cyclin-dependent kinases constitute the core components of the cell cycle clock#
- the cell cycle uses protein kinases to execute the various steps of cell cycle progression, e.g.:
- phosphorylation of centrosome-associated proteins allows for their duplication
- phosphorylation of nuclear membrane proteins (e.g. lamin, nucleoporins) leads to dissolution of the membrane
- all cell cycle kinases are called cyclin-dependent kinases (CDKs)
- CDKs are serine/threonine kinases
- cyclins are the regulatory proteins
- CDKs are activated (given activating phosphorylations) by CDK-activating kinases (CAKs))
- cyclins
- increase kinase activity of Cdks
- physically guide Cdks to substrates
- have cyclic pattern of concentration in cells (CDK concentrations don't vary)
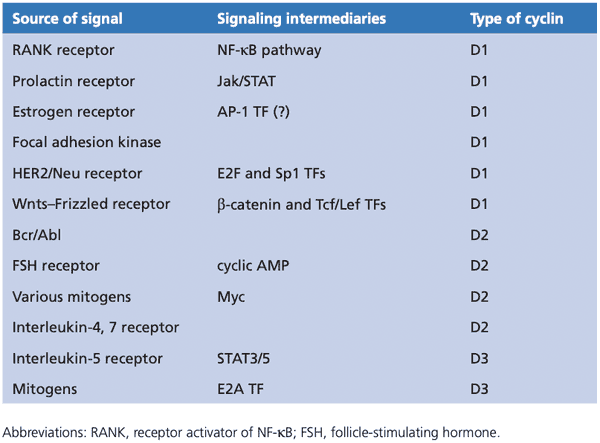
Cyclin-CDK pairings#
- Before R point in G1
- CDK4 and CDK6
- D-type cyclins D1, D2, D3
- After R point in G1
- CDK2
- E-type cyclins E1, E2
- Early S phase
- CDK2
- A-type cyclins A1, A2
- Late S phase
- CDC2/CDK1
- A-type cyclins A1, A2
- G2 phase
- CDC2/CDK1
- B-type cyclins B1, B2
- still not fully clear how later cyclin-CDK complexes suppress the activities of their predecessors
- fall in cyclin levels is degradation via ubiquitylation
-
D-type cyclins are an exception: they do NOT follow well-programmed fluctuations
- half life of cyclin D1 is ~30 minutes
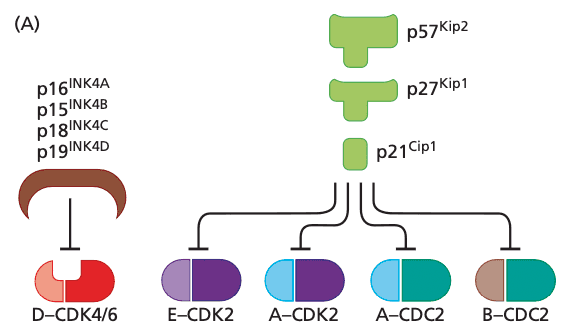
Cyclin-CDK complexes are also regulated by CDK inhibitors#
- seven known CDK inhibitors
- group of 4 CKIs are the INK4 proteins (inhibitors of CDK4)
- p16INK4A, p15INK4B, p18INK4C, p19INK4D
- p21Cip1/Waf1, p27Kip1, p57Kip2 are more widely acting and can inhibit all cyclin-CDK complexes after R point
- Cip/Kip CKIs also affect processes beyond cell cycle progression, e.g.
- transcriptional regulation
- apoptosis
- cell fate determinination
- cell migration
- cytoskeletal organization
- p57Kip2 mostly important in embryogenesis and not cancer
- Cip/Kip CKIs also affect processes beyond cell cycle progression, e.g.
- group of 4 CKIs are the INK4 proteins (inhibitors of CDK4)
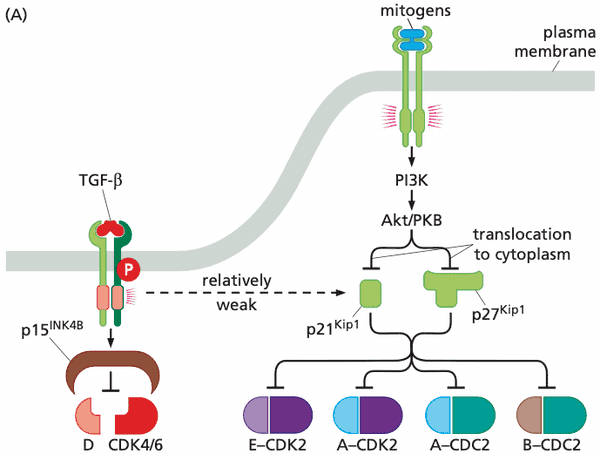
When CDK inhibitors are activated#
- TGF-β increases concentration of p15INK4B which inhibits D:CDK4/6, preventing advance through the R point
- DNA damage activates p21Cip1/Waf1 which prevents cell cycle advance until DNA is repaired
- p21Cip1/Waf1 also inhibits PCNA (proliferating-cell nuclear antigen), a required component of DNA replication DNA replication is halted
Mitogens override CDK inhibitors#
- Akt/PKB phosphorylates p21Cip1/Waf1 and p27Kip1 preventing them from entering the nucleus and inhibiting the cyclin-CDK complexes
- paradoxically, however, p21Cip1/Waf1 and p27Kip1 stimulates formation of cyclin D-CDK4/6 complexes
Post-mitotic state is likely imposed by CDK inhibitors#
- in mice cerebella, post-mitotic differentiated cells have high levels of p27Kip1
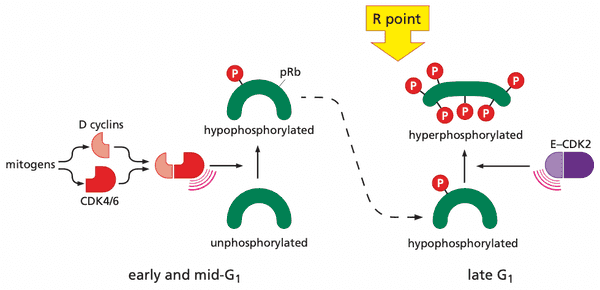
Viral oncoproteins reveal how pRb blocks advance through the cell cycle#
- pRb phosphorylation pattern
- none in G0
- low after entrance to G1
- high soon after advance through R point
- high throughout remainder of cell cycle
- low after exiting mitosis
- phosphates removed by protein phosphatase type 1 (PP1)
- three viral oncoproteins: E1A, large T antigen, E7
- structurally distinct
- all bind pRb
- evolution suggests that binding pRb improves viral replication
- viral oncoproteins only sequester hypophosphorylated pRb (the pRb state that inhibits cell cycle)
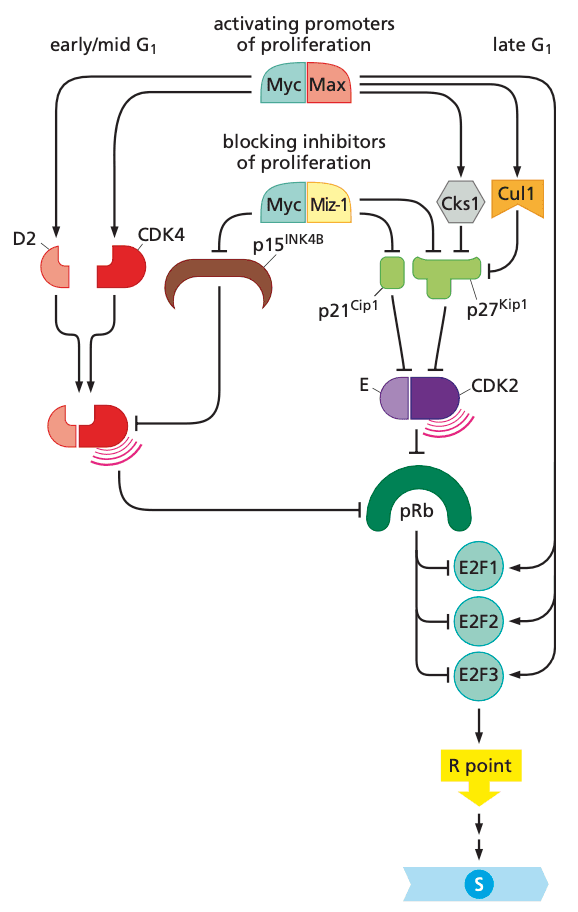
pRb is deployed by the cell cycle clock to serve as a guardian of the restriction-point gate#
-
(hypo)phosphorylation by cyclin D-CDK4/6 is necessary for cyclin E-CDK2 to bind and perform its kinase function
- more cyclin D-CDK4/6 = more likely that cyclin E-CDK2 hyperphosphorylate pRb
- cyclin D-CDK4/6 also sequester p27Kip1, liberating cyclin E-CDK2 to start hyperphosphorylation
- pocket of pRb, p107, and p130 bind E2F transcription factors
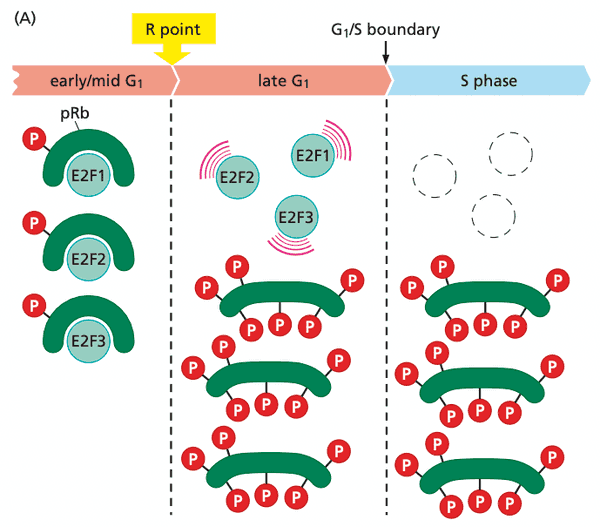
E2F transcription factors enable pRb to implement growth-versus-quiescence decisions#
- pRb suppresses G1 advance by acting on E2F transcription factors
- hypophosphorylated pRb: binds E2Fs
- hyperphosphorylated pRb: dissociates from E2Fs
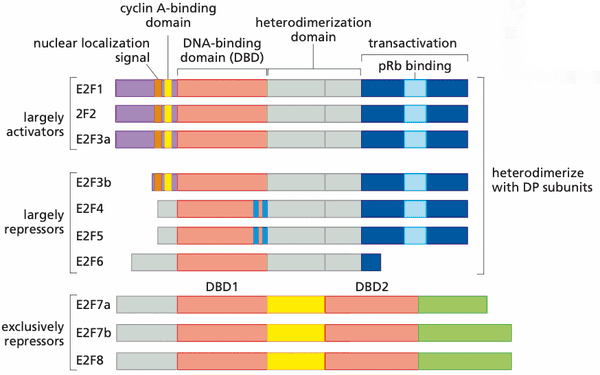
- E2F 1 through 6 form heterodimers with DP1 or DP2
- E2F 7 and 8 have two DNA binding domains (and therefore don't need to dimerize)
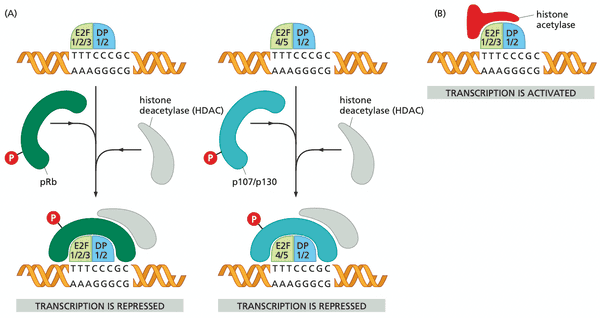
- pRb blocks transactivation domain of E2F and recruits HDAC for chromatin remodeling.
- E2Fs activate ~500 genes
- E2Fs activate positive-feedback loop to drive hyperphosphorylation of pRb to completion
A variety of mitogenic signaling pathways control the phosphorylation state of pRb#
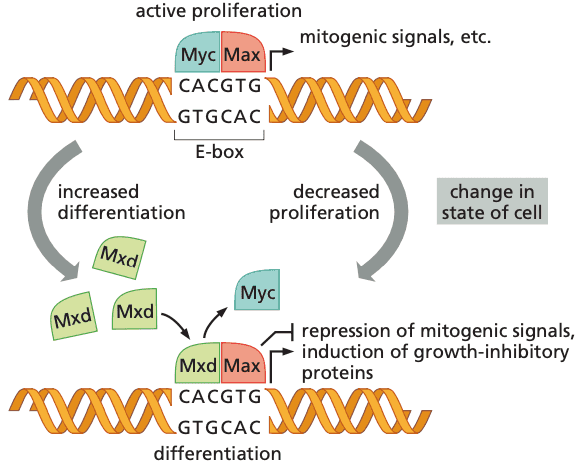
The Myc protein governs decisions to proliferate or differentiate#
- many tumors express c-Myc, or its neighbors N-Myc and L-Myc.
- Myc is found in the nucleus and not as signaling proteins in the cytoplasm (like Ras and Src).
- bHLH transcription factors bind to E-boxes (enhancer box) (which have the sequence CACGTG).
- Myc-Max = proliferation
- allows RNApolII to transcribe full-length mRNA transcripts
- Mxd-Max = differentiation
- prevents RNApolII from transcription
-
Myc's effects on the cell cycle all promote progress through the G1 phase
- promotes transcription of cyclin D2
- activates Cks1 and Cul1 which stop p27Kip1 from inhibiting cyclin E-CDK2, enabling cyclin E-CDK2 to hyperphosphorylate pRb
TGF-β prevents phosphorylation of pRb and thereby blocks cell cycle progression#
pRb function and the controls of differentiation are closely linked#
Control of pRb function is perturbed in most if not all human cancers#
12, 13. Metabolism in cancer#
- Readings
- Weinberg, The Biology of Cancer 2e
- pp. 53-55
- pp. 265-268
- Weinberg, The Biology of Cancer 2e
Cancer cells exhibit an altered energy metabolism @weinberg2e/2.6#
Von Hippel-Lindau disease: pVHL modulates the hypoxic response @weinberg2e/7.12#
- VHL is a tumor suppressor gene
- pVHL (protein product of VHL gene) promotes degradation of HIF-2 transcription factor (HIF-1 is the protein characterized experimentally, however)
- in normoxia conditions (detected by proline hydroxylase)
- pVHL promotes degradation of HIF-1 subunit cell only has low levels of HIF-1
- HIF-1 induces expression of VEGF, PDGF, and TGF-α.
- more than 200 genes induced by HIFs
14, 15. Cell death, senescence, and cancer @weinberg2e/9#
- Readings
- Weinberg, The Biology of Cancer 2e: pp. 331-381
Papovaviruses lead to the discovery of p53#
p53 is discovered to be a tumor suppressor gene#
Mutant versions of p53 interfere with normal p53 function#
p53 protein molecules usually have short lifetimes#
A variety of signals cause p53 induction#
DNA damage and deregulated growth signals cause p53 stabilization#
Mdm2 destroys its own creator#
ARF and p53-mediated apoptosis protect against cancer by monitoring intracellular signaling#
p53 functions as a transcription factor that halts cell cycle advance in response to DNA damage and attempts to aid in the repair process#
p53 often ushers in the apoptotic death program#
p53 inactivation provides advantage to incipient cancer cells at a number of steps in tumor progression#
Inherited mutant alleles affecting the p53 pathway predispose one to a variety of tumors#
Apoptosis is a complex program that often depends on mitochondria#
Both intrinsic and extrinsic apoptotic programs can lead to cell death#
Cancer cells invent numerous ways to inactivate some or all of the apoptotic machinery#
Necrosis and autophagy: two additional forks in the road of tumor progression#
16. Stem cells and cancer stem cells#
- Readings
- Weinberg, The Biology of Cancer 2e
- pp. 458-463
- pp. 206-209
- pp. 259-265
- Weinberg, The Biology of Cancer 2e
Tumor stem cells further complicate the Darwinian model of clonal succession and tumor progression @weinberg2e/11.6#
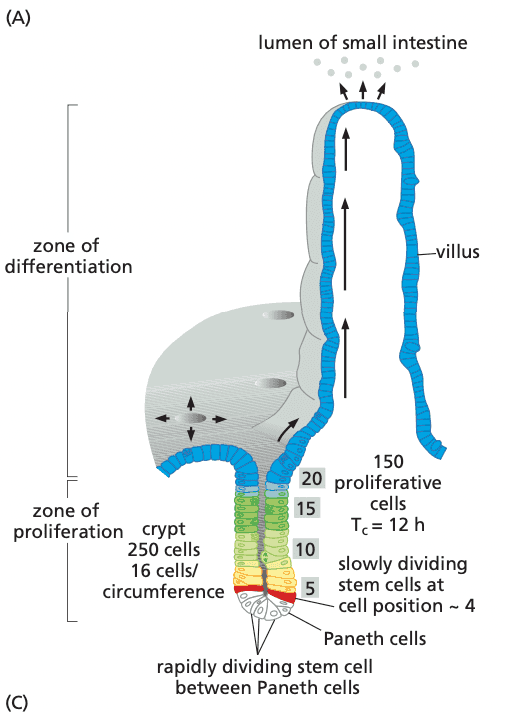
Apc facilitates egress of cells from colonic crypts @weinberg2e/7.11#
17. The tumor microenvironment @weinberg2e/13#
- Readings
- Weinberg, The Biology of Cancer 2e, pp. 577-639
Normal and neoplastic epithelial tissues are formed from interdependent cell types#
- diverse stromal cell types within tumors are all members of several mesenchymal cell lineages that generate both connective tissue and various types of immune cells
- heterotypic signaling. signaling between dissimilar cell types
- types of signals
- mitogenic growth factors (hepatocyte growth factor (HGF), transforming growth factor alpha (TGF), platelet-derived growth factor (PDGF)).
- growth-inhibitory signals (transforming growth factor beta (TGF)).
- trophic factors (insulin-like growth factor 1, 2 (IGF1, IGF2)).
- stromal-epithelial interactions play key role in tumor progression (including tumor formation)
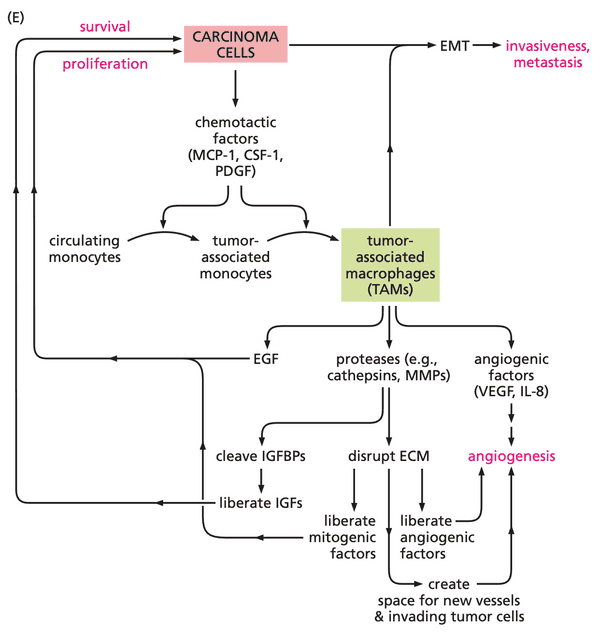
- carcinoma cells
- release growth factors, cytokines, chemokines; and
- recruit macrophages, neutrophils, and lymphocytes
- release TNF- and prostaglandins inflammatory response proliferation of nearby epithelial cells + angiogenesis
- carcinoma cells
- examples of heterotypic interactions
- carcinoma: PDGF fibroblasts, myofibroblasts, macrophages IGF-1
- melanoma: PDGF fibroblasts IGF-2 production
- breast cancer: stromal cells release SDF-1/CXCL12 (chemokine) + HGF/SF (growth factor) proliferation and survival of nearby epithelial cancer cells
- anoikis. a form of apoptosis resulting from loss of anchorage to a solid substrate
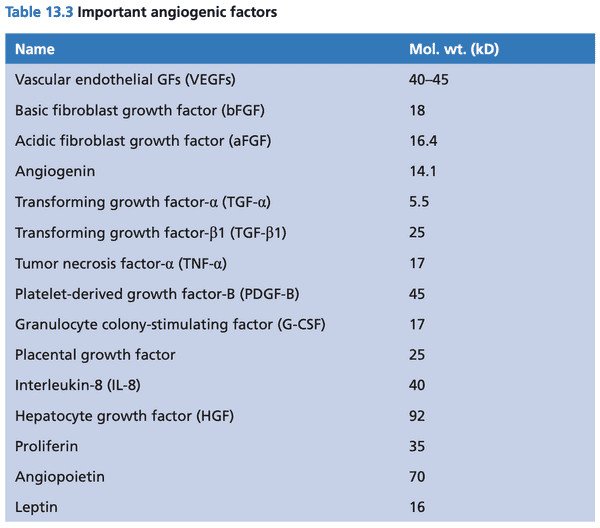
- angiogenic factors stimulate growth of capillaries (endothelial cells)
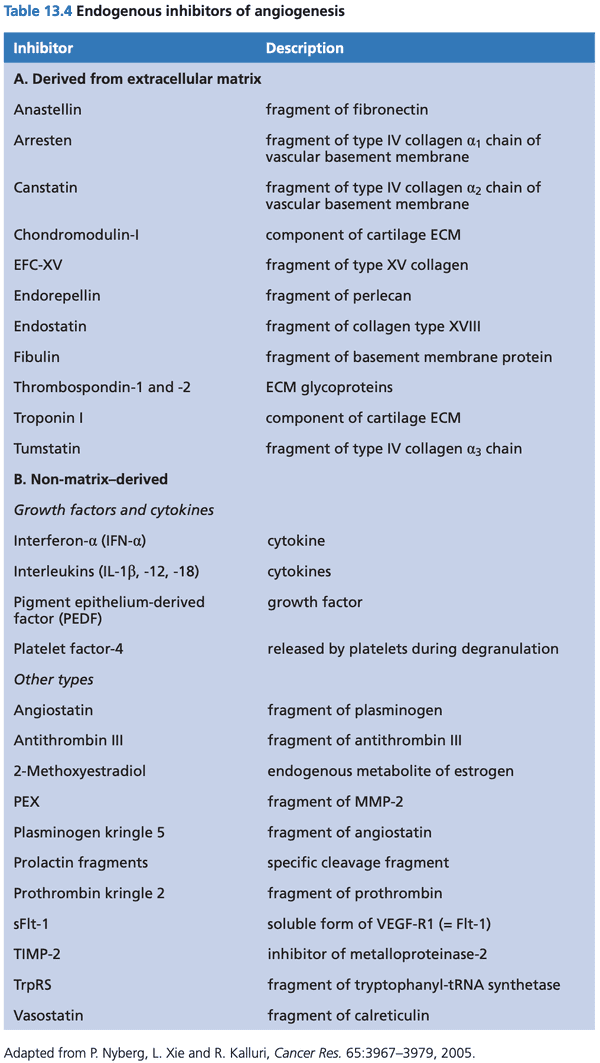
- endothelial cells stimulate growth of pericytes and vascular smooth muscle cells (collectively called mural cells) by releasing PDGF and HB-EGF (heparin-binding EGF).
- pericytes release VEGF and Ang-1 (angiopoietin-1)
- endothelial cells stimulate growth of pericytes and vascular smooth muscle cells (collectively called mural cells) by releasing PDGF and HB-EGF (heparin-binding EGF).
- ascitic tumors. tumors that are almost totally independent and can accumulate in various body fluids
The cells forming cancer cell lines develop without heterotypic interactions and deviate from the behavior of cells within human tumors#
- cancer cell lines are not always predictive of actual drug efficacy because stroma is not present; usually stromal cells can survive in tissue cultures
- epithelial cells that can survive in vitro are selected for; not representative of the tumor's true population
- PDX (patient derived xenografts) in immunocompromised mice is not necessarily predictive of eventual clinical responses to drugs
- can be due to ectopic vs. orthotopic
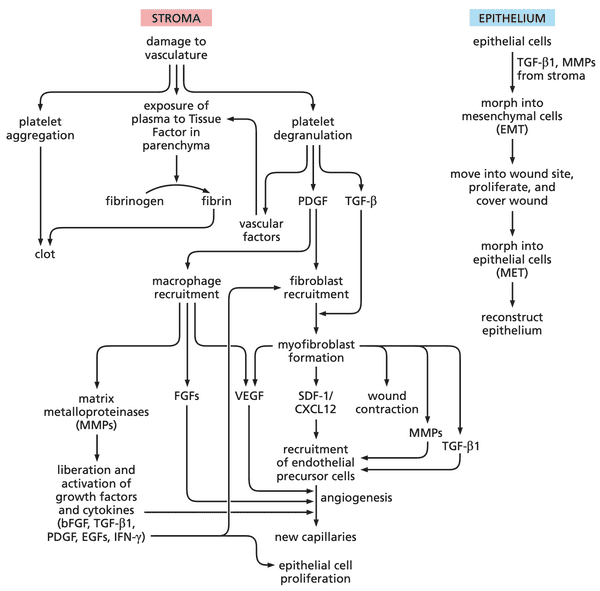
Tumors resemble wounded tissues that do not heal#
- cancer cells co-opt the wound healing program
- platelets release PDGF and TGF-
- wounding causes release of vasoactive factors (increases permeability of blood vessels near wound)
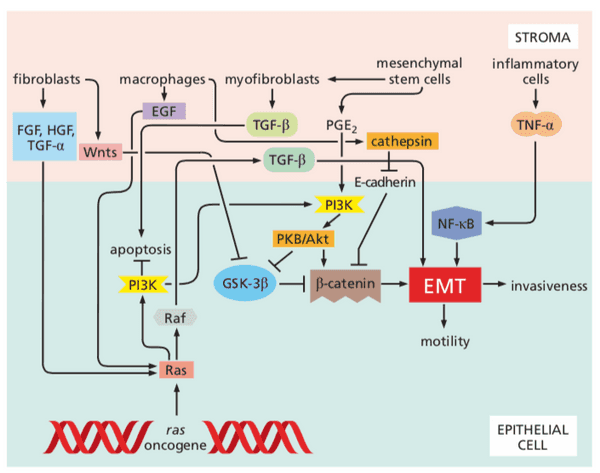
- PDGF attracts fibroblasts TGF- converts fibroblasts to myofibroblasts myofibroblasts release matrix metalloproteinases (MMPs)
- MMPs carry a zinc ion to carry out catalysis
- 23 MMPs have been documented in mammalian cells
- carves out space for new cells
- releases growth factors embedded in ECM: basic FGF (bFGF), TGF-1, PDGF, EGF-related factors, and IFN-.
- fibroblasts also release various fibroblast growth factors (FGFs)
- immune cells recruited: monocytes ( macrophages), neutrophils, eosinophils, mast cells, lymphocytes
- release VEGF
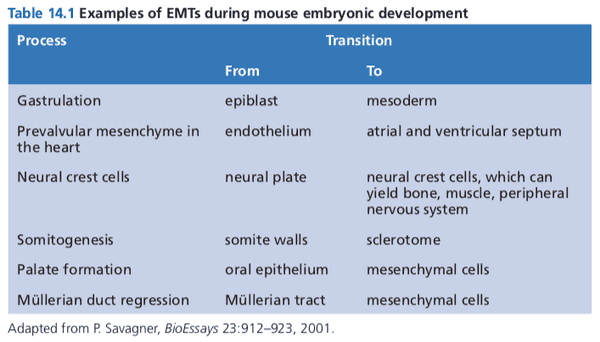
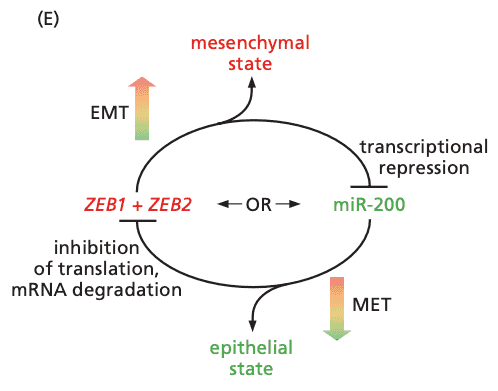
- epithelial cells undergo EMT to become motile and invasive
- once wound healing is complete, cells undergo MET to become epithelial again
- evidence of parallels:
- presence of clumps of fibrin in the tumor-associated stroma
- due to leakiness of newly formed blood vessels, rather than traumatic damage
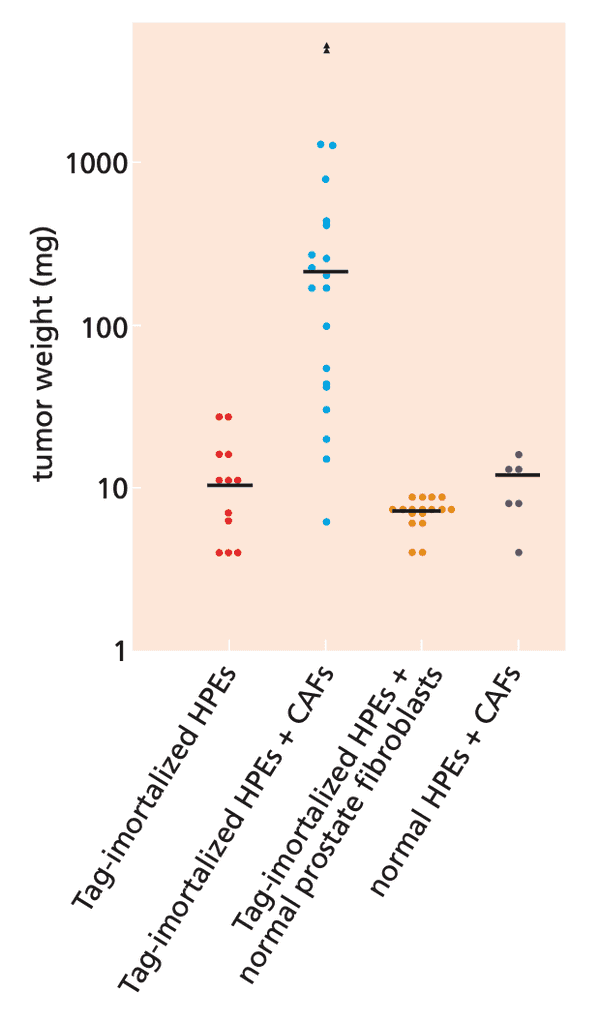
- many kinds of cancer cells (e.g. breast, prostate, colon, and lung carcinomas) continuously release PDGF
- initial scaffold: fibrin
- collagen forms the later, more permanent matrix
- myofibroblasts perform the physical contraction to close a wound
- in prostate carcinomas, we can identify myofibroblasts because myofibroblasts produce vimentin, while the normal tissue does not.
- @research where do stromal fibroblasts and myofibroblasts originate?
- existing fibroblasts can transform into myofibroblasts upon contact with TGF-1
- fibrocytes and mesenchymal stem cells
- tumor stroma is desmoplastic (more ECM produced by myofibroblasts) compared to normal tissue stroma
- collagen I and III
- fibronectin
- proteoglycans
- glycosaminoglycans (GAGs)
- secrete urokinase plasminogen activator (uPA) and MMPs
- ECM eventually becomes acellular and collagenous
- carcinoma-associated fibroblasts. describes the mixed populations of fibroblasts and myofibroblasts present in the stroma of epithelial tumors
Experiments directly demonstrate that stromal cells are active contributors to tumorigenesis#
- keratinocytes transfected with PDGF expression vector did not proliferation in vitro; when implanted into host mice, suddenly became tumorigenic because stromal cells had PDGF receptors
- stromal cells then drove proliferation of PDGF-secreting keratinocytes
- carcinoma-associated fibroblasts (CAFs) are much different than normal fibroblasts
- mechanism still not entirely known
- CAF myofibroblasts release SDF-1/CXCL12 (stroma-derived factor 1) that recruits circulating EPCs (endothelial precursor cells, come from bone marrow) + other myeloid cell types leads to formation of neovasculature
- neovasculature assumed to be formed by existing endothelial cells and EPCs
- discovered that cancer cells can transdifferentiate into endothelial-like cells
Macrophages and myeloid cells play important roles in activating the tumor-associated stroma#
- chemotactic factors. chemical factors that provide directional cues
- chemokines = chemotactic cytokines
- MCP-1 (monocyte chemotactic protein-1). attracts monocytes to tumors and induce differentiation into macrophages
- CSF-1 (M-CSF, macrophage colony-stimulating factor). stimulates monocyte-to-macrophage differentiation
- macrophages stimulate angiogenesis
- macrophages secrete VEGF and IL-8
- macrophages appear quite able to tolerate hypoxia
- macrophages release MMP-9, which cleaves IGFBPs and liberates IGF-1, a survival signal
- a second myeloid lineage cell type involved in tumor invasiveness: immature myeloid cells (IMCs)
- cancer cells secrete CCL9, which recruits IMCs
- IMCs secrete MMP-2 and MMP-9 (needed for invasion and metastasis)
Endothelial cells and the vessels that they form ensure tumors adequate access to the circulation#
- 0.2mm is the distance that oxygen can effectively diffuse through living tissue
- production of VEGF is governed by availability of oxygen
- cells use VHL complex to sense intracellular oxygen tension
- HIF-1 and HIF-1 transcription factors accumulate express genes to encourage angiogenesis, e.g. VEGF
- similar pathway occurs in low pH conditions
- cells use VHL complex to sense intracellular oxygen tension
- two VEGF receptors: VEGFR-1/Flt-1 and VEGFR-2/Flk-1/KDR + bFGF receptor
- found on endothelial cell membranes
- results in endothelial cell proliferation and morphological changes to form capillary walls
- initial capillaries formed from EPCs; later capillaries formed with already present endothelial cells in tumor stroma
- similar mechanisms operate during the formation of lymphatics: lymphangiogenesis
- VEGF-C and VEGF-D are the relevant growth factors
- factors other than VEGF involved in angiogenesis: TGF-, basic fibroblast growth factors (notably FGF2), IL-8, angiopoietin, angiogenin, PDGF
- pericytes are tightly attached to endothelial cells in normal vessels; they are loosely attached in tumors
- may be due to imbalance of Ang-1 and Ang-2 (antagonistic angiopoietins) and elevated levels of VEGF-A
- endothelial cells in tumor masses often leave gaps among themselves, resulting in leaky vessels
- causes of high hydrostatic pressure in tumors
- lymphatic vessel collapse
- leaky capillaries
- fibroblast contraction (due to high levels of PDGF)
Tripping the angiogenic switch is esssential for tumor expansion#
- the ability to attract blood vessels is not automatic
- dynamics of angiogenic switch stem from studies of the Rip-Tag transgenic mouse
- expression of SV40 large and small T antigen is driven by promoter of insulin gene
- useful because endocrine cells are easily distinguished from the surrounding exocrine cells
- 8 to 12% of hyperplastic islets become angiogenic
- VEGF is generally sequestered by the ECM
- when inflammatory cells (mast cells and macrophages) release MMP-9 and degrade the ECM, VEGF is freed and can induce angiogenesis
- angiogenic switch (in this tissue) depends on ability to recruit inflammatory cells
- Rip-Tag model does not typify all kinds of angiogenic switches
- VEGF-A/B, a/bFGF, TGF- are all angiogenic factors (angiogenic switch uses different combinations in different cancers)
The angiogenic switch initiates a highly complex process#
- angiogenesis begins in stroma before basement membrane broken down (angiogenic signals travel through porous basement membrane)
- angiogenesis speeds up greatly only when cancer cells become invasive
- myofibroblasts also foster angiogenesis through release of SDF-1/CXCL12, which recruits EPCs to the tumor
- Id genes encode TFs regulating differentiation of a variety of cell types
- mice with EPCs knocked out (using Id gene knockouts) don't have tumors progress until wild-type stem cells are transplanted back into the mice
Angiogenesis is normally suppressed by physiologic inhibitors#
- plots of tumor volume vs. microvessel density suggest that angiogenesis acts as a rate-limiting determinant of tumor growth
- angiogenesis burst usually stopped quickly
- in healthy tissue, HIF-1 TF only assembles under hypoxic conditions
- thombospondin-1 (Tsp-1) protein binds to CD36 receptor on endothelial cells and halts their proliferation
- also leads endothelial cells to release FasL, a pro-apoptotic signaling protein
- Fas receptor displayed on newly formed endothelial cells, and not on mature cells
- TSP1 transcription is strongly induced by p53
- Ras shuts down Tsp-1 production
- other inhibitors exist
- often, excision of tumor can also excise the inhibitors and increase production of wound-healing growth factors, which leads to development of already seeded metastases
- inhibitors include angiostatin and endostatin
- MMP antagonists called tissue inhibitors of metalloproteinases (TIMPs)
- @research TIMP MoA still not elucidated
- often, excision of tumor can also excise the inhibitors and increase production of wound-healing growth factors, which leads to development of already seeded metastases
Anti-angiogenesis (AAG) therapies can be employed to treat cancer#
- most AAG therapies target normal cells that have been co-opted by cancers
- these cells are usually cycling and not quiescent, which makes them more vulnerable to cytotoxic therapies
- endostatin and angiostatin + chemotherapy drug combination appears to work
- IFN- and IFN-. another class of natural AG inhibitors
- AAG agents when blocking multiple pathways (possibly on different cell types) likely can starve a tumor of blood
- alone, can only normalize vasculature
- VEGF important in nascent tumors; PDGF important in advanced tumors
18. Metastasis @weinberg2e/14#
- Readings
- Weinberg, The Biology of Cancer 2e, pp. 641-720
Travel of cancer cells from a primary tumor to a site of potential metastasis depends on a series of complex biological steps#
- local invasion depends invariably on the release of secreted proteases, such as MMP-2 and MMP-9
- some cancer cells make their own proteases
- some cancers co-opt stromal cells to release these enzymes
- process of intravasation is less well studied
- in breast cancers, triad of caricnoma cells, macrophages, and endothelial cells assembles to enable intravasation
- macrophages release EGF that stimulate cancer cells to invade
- cancers with high densities of these triads predicts eventual metastatic relapse
- incomplete understanding of cancer cells in general circulation (circulating tumor cells, or CTCs)
- cancer cells and cancer cell-platelet aggregates are much larger than capillary diameter within minutes of entering venous circulation many cancer cells will lodge in the capillary beds of the lungs
- these cells may then pinch off cytoplasm, freeing them to travel to other capillary beds (since mets exist beyond the lung)
- cancers may also co-opt functions of macrophages to extravasate
Colonization represents the most complex and challenging step of the invasion-metastasis cascade#
- colonization. the growth from microscopic metastases into macroscopic metastases
- detecting micromets
- anti-cytokeratins for micromets in bone marrow and blood
- antiEpCAM useful for micromets in lymph nodes
- metastatic shower
- the second wave of mets that arise after the first mets successfully colonize are much more deadly since they inherit the colonization phenotype
- however, mets in terminal cancer patients are generally genetically diverse; do the second wave of mets diversify, or did many diverse mets undergo secondary showers?
THe epithelial-mesenchymal transition and associated loss of E-cadherin expression enable carcinoma cells to become invasive#
- EMT in normal physiology
- part of wound healing
- part of gastrulation, when cells leave the ectoderm to form the mesoderm
- E-cadherin, which forms adherens junctions, is pivotal to epithelial phenotype
- suppressing expression of E-cadherin leads to acquisition of mesenchymal morphology and increased motility
- E-cadherin is replaced by N-cadherin during EMT
- HGF (hepatocyte growth factor) promotes E>N cadherin transition
- Because stromal cells express N-cadherin, expression of N-cadherin by cancer cells helps them insert themselves into the stroma (invasion).
- N-cadherin forms weaker intermolecular bonds, thus facilitating motility as opposed to E-cadherin
Epithelial-mesenchymal transitions are often induced by contextual signals#
- expression of integrin is associated with the EMT
- micromets undergo MET likely because new tumor microevironment has not yet been colonized and no EMT growth factors are present (yet)
- mesenchymal cancer cells don't proliferate well; a MET is necessary for outgrowth of metastatic colonies
- two possibilities for activating EMTs
- by the stroma, providing heterotypic signals to the tumor (most evidence points to this cause)
- by the tumor cells, who sense that they are not surrounded by other epithelial cells
- stromal signals include
- Wnts (both canonical and non-canonical)
- TGF
- TNF
- EGF (epidermal growth factor)
- HGF (hepatocyte growth factor)
- IGF1
- PGE2 (prostaglandin E2)
- genetic alterations in carcinoma cells further increase their responsiveness to these signals
- TGF is typically anti-proliferative
- most carcinomas lose the pRb pathway that executives TGF's cytostatic effects
- TGF and canonical Wnt proteins are secreted by epithelial cells normally, but ability to activate EMT is blocked by secreted inhibitory proteins as well: BMPs, SFRP1, DKK1
Stromal cells contribute to the induction of invasiveness#
- CSF-1 and TAMs contribute to invasive growth
- CSF-1 knockout = no TAMs recruited
- primary tumors still grow, but do not show invasive phenotype
- molecular signals produced by macrophages still not clear
- likely contributors (not complete)
- macrophage-derived TNF (initial invasion)
- EGF (during intravasation)
- macrophages can release cathepsin B protease that may degrade E-cadherin and lead to EMT
- likely contributors (not complete)
EMTs are programmed by transcription factors that orchestrate key steps of embryogenesis#
- there are likely many kinds of EMTs
- cancers may only activate parts of an EMT
EMT-inducing transcription factors also enable entrance into the stem cell state#
- tumor cells need stem cell state to seed new tumors
- this stem cell state can be entered via EMT-inducing TFs
- evidence that EMT-TFs engender SC-like abilities
- mammary epithelial cells (MECs) without exposure to Slug and Sox9 unable to generate a mammary ductal tree
- mammary epithelial cells (MECs) with exposure to Slug and Sox9 100x able to generate a mammary ductal tree
- myofibroblasts produce PGE2
- PGE2
- induces entrance of epithelial cells into SC compartment
- may facilitate Wnt signaling
- PGE2
- unlikely (why?) that EMTs help micromets colonize into macromets (i.e. EMTs probably do not help cancer cells adapt to foreign microenvironments)
EMT-inducing TFs help drive malignant progression#
- ZEB1 and ZEB2 EMT-TFs determine whether cells stay in epithelial or mesenchymal state (a bistable switch)
- ZEB1 + ZEB2 compete with miR-200 microRNAs
- shutting down actions of EMT-TFs in in vivo tumors decreased number of metastases by ~85%; the remaining were due to tumor cells that did not have Twist (an EMT-TF) expression shut off
- shutdown of Twist enabled faster growth of primary tumor; therefore prevention of metastases cannot be attributed to a simple cytostatic effect
- Twist and Slug enable cells to resist apoptosis and anoikis
- has not been shown systematically yet however
- many factors involved in tumor microenvironment also are involved in metastasis; EMT may be a natural consequence of a growing tumor
- summary
- many malignant cell phenotypes are induced by heterotypic signals from the stroma (non-genetic changes)
- because EMT signals come from the stroma, carcinoma cells may revert back to epithelial upon leaving the primary tumor
- EMT-TFs often express together, leading to possibility that many malignant traits are acquired simultaneously
Extracellular proteases play key roles in invasiveness#
- matrix metalloproteinases (MMPs) allow cancer cells to excavate passageways through the ECM and remodel the nearby tissue environment
- most MMPs are secreted by recruited stromal cells rather than the tumor cells themselves
- macrophages
- neutrophils
- fibroblasts
- MMPs can cleave
- fibronectin
- tenascin
- laminin
- collagens
- proteoglycans
- and liberate growth factors that were sequestered in the ECM
- cancer cells express MT1-MMP (membrane type-1 MMP) on the surface of their plasma membranes
- MT1-MMP can cleave pro-enzymes (e.g. pro-MMP-2) into active enzymes
- cell nuclei are relatively rigid and cannot be compressed
- MT1-MMP are concentrated at cancer cell invadopodia
-
the remodeling of ECM takes place continuously in mitotically active tissues
- clinical trials targeting MMP were terminated because of effects on remodeling of cartilage and joints (led to high levels of joint stiffness and pain)
- MMPs are negatively regulated by tissue inhibitors of metalloproteinases (TIMPs)
- MMPs alone can allow cancer cells to progress through all stages of multi-step tumorigenesis
Small Ras-like GTPases control cellular processes such as adhesion, cell shape, and cell motility#
Metastasizing cells can use lymphatic vessels to disperse from the primary tumor#
A variety of factors govern the organ sites in which disseminated cancer cells form metastases#
Metastasis to bone requires the subversion of osteoblasts and osteoclasts#
Metastasis supressor genes contribute to regulating the metastatic phenotype#
Occult micrometastases threaten the long-term survival of cancer patients#
19, 20. Cancer immunology and immunotherapy @weinberg2e/15#
- Readings
- Weinberg, The Biology of Cancer 2e, pp. 723-796
The immune system functions to destroy foresign invaders and abnormal cells in the body's tissues#
The adaptive immune response leads to antibody production#
Another adaptive immune repsonse leads to the formation of cytotoxic cells#
The innate immune response does not require prior sensitization#
The need to distinguish self from non-self results in immune tolerance#
Regulatory T cells are able to suppress major components of the adaptive immune response#
The immunosurveillance theory is born and then suffers major setbacks#
Use of genetically altered mice leads to a resurrection of the immunosurveillance theory#
The human immune system plays a critical role in warding off various types of human cancer#
Subtle differences between normal and neoplastic tissues may allow the immune system to distinguish between them#
Tumor transplantation antigens often provoke potent immune responses#
Tumor-associated transplantation antigens may also evoke anti-tumor immunity#
Cancer cells can evade immune detection by suppressing cell-surface display of tumor antigens#
Cancer cells protect themselves from destruction by NK cells and macrophages#
Tumor cells launch counterattacks on immunocytes#
Cancer cells become intrinsically resistant to various forms of killing used by the immune system#
Cancer cells attract regulatory T cells to fend off attacks by other lymphocytes#
Passive immunization with monoclonal antibodies can be used to kill breast cancer cells#
Passive immunization with antibody can also be used to treat B-cell tumors#
Transfer of foreign immunocytes can lead to cures of certain hematopoietic malignancies#
Patients' immune systems can be mobilized to attack their tumors#
21, 22. Principles of cancer therapy @weinberg2e/16#
- Readings
- Weinberg, The Biology of Cancer 2e, pp. 797-875
The development and clinical use of effective therapies will depend on accurate diagnosis of disease#
Surgery, radiotherapy, and chemotherapy are the major pillars on which current cancer therapies rest#
Differentiation, apoptosis, and cell cycle checkpoints can be exploited to kill cancer cells#
Functional considerations dictate that only a subset of the defective proteins in cancer cells are attractive targets for drug development#
The biochemistry of proteins also determines whether they are attractive targets for intervention#
Pharmaceutical chemists can generate and explore the biochemical properties of a wide array of potential drugs#
Drug candidates must be tested on cell models as an initial measurement of their utility in whole organisms#
Studies of a drug's action in laboratory animals are an essential part of pre-clinical testing#
Promising candidate drugs are subjected to rigorous clinical tests in Phase I trials in humans#
Phase II and III trials provide credible indications of clinical efficacy#
Tumors often develop resistance to initially effective therapy#
Gleevec paved the way for the development of many other highly targeted compounds#
EGF receptor antagonists may be useful for treating a wide variety of tumor types#
Proteasome inhibitors yield unexpected therapeutic benefit#
A sheep teratogen may be useful as a highly potent anti-cancer drug#
mTOR, a master regulator of cell physiology, represents an attractive target for anti-cancer therapy#
B-Raf discoveries have led to inroads into the melanoma problem#
L21, L22#
What is the goal of treatment?#
- live longer (decreased mortality), live better (decreased morbidity)
- remember: the goals of care matter
- we are treating a patient, not a tumor
How does cancer create health problems? Why does it kill?#
- tumors can alter normal organ function
- mass effect (blockage). the presence of a mass blocks organ function
- invade normal structures. invade normal structures that disrupt function e.g. brain
- produce hormones that change physiology
- endocrine: seratonin, insulin
- nonendocrine: parathyroid high
- paraneoplastic syndrome
- a group of rare disorders triggered by an abnormal immune response to a cancerous tumor
- weight loss; altered systemic metabolism
- cachexia. loss of mean body mass despite adequate nutrition
- infection
- loss of immune cells
- potential causes: leukemia, malnutrition, systemic therapy
- loss of barrier function
- loss of immune cells
- coagulopathies
- bleeding
- loss of vascular function
- clotting
- can be due to liver dysfunction (release tissue factor in response to vascular damage)
- leads to pulmonary embolism, disseminated intravascular coagulation (DIC)
- bleeding
- dehydration
- kidney dysfunctions
- malnutrition
- pain
How is cancer treated?#
- local therapy
- surgery
- radiation
- cryotherapy
- radio waves
- electric fields
- systemic therapy
- chemotherapy
- antibodies
- cell therapies
- vitamins
- hormones
- vaccines
Local therapy#
- Surgery
- major advance in 1846: anesthesia
- allowed for complex procedures
- identification and removal of surgical margins
- examination of lymph nodes
- major advance in 1846: anesthesia
- Radiation
- brachytherapy. close up
- used to treat cervical, prostate, brain cancer
- dose decays exponentially with distance
- external beam therapy.
-
- used to treat thyroid cancer (since thryoid metabolizes iodine)
-
- concentrates in bone (like calcium)
- -CD20
- direct radiation via antibody to a specific site
- total body radiation: to destroy immune system and blood cancers
- whole brain radiation: broad metastases to the brain
- stereotactic radiation. delivering external beam radiation to very specific sites
-
- brachytherapy. close up
Systemic therapy#
- why systemic therapy?
- best breast cancer imaging: ~0.5mm = 1 million cells
- thus we cannot see populations smaller than 1 million cells
- best breast cancer imaging: ~0.5mm = 1 million cells
- characteristics of systemic therapy
- (generally) informed by the molecular characteristics of the tumors
- pharmacology is critical
- sometimes very effective
- blood cancers (lymphomas), testicular cancer
- but many cancers do not respond
23. Therapy and resistance; early detection and prevention#
- Readings: TBD
@weinberg2e/10 Eternal life: cell immortalization and tumorigenesis#
Normal cell populations register the number of cell generations separating them from their ancesters in the early embryo#
Cancer cells need to become immortal in order to form tumors#
Cell-physiologic stresses impose a limitation on replication#
The proliferation of cultured cells is also limited by the telomeres of their chromosomes#
Telomeres are complex molecular structures that are not easily replicated#
Incipient cancer cells can escape crisis by expressing telomerase#
Telomerase plays a key role in the proliferation of human cancer cells#
Some immortalized cells can maintain telomeres without telomerase#
Telomeres play different roles in the cells of laboratory mice and in human cells#
Telomerase-negative mice show both decreased and increased cancer susceptibility#
The mechanisms underlying cancer pathogenesis in telomerase-negative mice may also operate during the development of human tumors#
@weinberg2e/11 Multi-step tumorigenesis#
Histopathology provides evidence of multi-step tumor formation#
- biopsies reveal a variety of tissue states with differing degrees of abnormality
- evidence for progression rather than leapfrogging/dead ends
- removal of polyps (dysplasia and hyperplasia) result in 80% reduction in incidence of colon carcinomas
Cells accumulate genetic and epigenetic alterations as tumor progression proceeds#
- later histological stage more genetic and epigenetic alterations
- genetic loci identities
- 5q21: APC
- 17p13: p53 tumor suppressor gene
- 18q: still unknown
Multi-step tumor progression helps to explain familial polyposis and field cancerization#
- family polyposis can be explained by multi-step tumor progression
- the first step is inherited through the germ line, which increases the likelihood of developing polyps
- multi-step tumor progression can also explain field cancerization
- normal phenotype but genetically damaged cells can proliferate in a patch
- later, two cells have separate mutations that enable them to form tumors
- without multi-step, probability to two tumors occuring independently is very unlikely
Cancer development seems to follow the rules of Darwinian evolution#
- the one distinction is that epigenetic factors play a role in cancer evolution, but epigenetic factors have not been show to play a role in species evolution
Tumor stem cells further complicate the Darwinian model of clonal succession and tumor progression#
- experiments used FACS to sort cells based on cell surface proteins (clusters of differentiation)
- if CSCs don't replicate frequently, how do they acquire a large number of mutations?
- may be that non-SC (transit-amplifying cells, =TA cells) become CSCs
A linear path of clonal succession oversimplifies the reality of cancer: intra-tumor heterogeneity#
- as tumors grows, the rate of genetic change outpaces Darwinian selection (and thus become heterogeneic)
- some colonies may grow symbiotically
- the heterogenetic and number of genetic alterations is seen in the adenoma stage, meaning that mutations occur relatively early in tumor development
The Darwinian model of tumor development is difficult to validate experimentally#
- functional genomics (looking at methylation) cannot distinguish between normal gene silencing and aberrant promoter methylation
- number of genetic alterations greatly exceeds the number of clonal successions
Multiple lines of evidence reveal that normal cells are resistant to transformation by a single mutated gene#
- transforming primary cells with ras oncogene does not transform them
- highlights biological difference between cell lines and primary cells
Transformation usually requires collaboration between two or more mutant genes#
- some viruses carry two oncogenes
- e.g. Polyomavirus carries middle T and large T
- large T: aid in adaptation to tissue culture conditions
- middle T: elicited phenotypes associated with ras oncogene
- rounding up of cells
- loss of contact inhibition
- acquisition of anchorage-independent growth
- ras and myc discovered to collaborate to transform cells
- transformed early-passage rat embryo fibroblasts and hamster kidney cells
- Ras-like oncoproteins are components of the cytoplasmic mitogenic signaling cascade
- Myc-like oncoproteins perturb the cell cycle control machinery, and operate in the nucleus